
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 656,603 – 656,655 |
| Length | 52 |
| Max. P | 0.736690 |

| Location | 656,603 – 656,655 |
|---|---|
| Length | 52 |
| Sequences | 5 |
| Columns | 57 |
| Reading direction | reverse |
| Mean pairwise identity | 56.31 |
| Shannon entropy | 0.78095 |
| G+C content | 0.46120 |
| Mean single sequence MFE | -9.62 |
| Consensus MFE | -2.58 |
| Energy contribution | -2.94 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.736690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 656603 52 - 21146708 ----UAGCCUACUACAAUUCGACCAUCCUGACAUUU-AAGAUCACCUGAUCUUGUCG ----.........................((((...-.(((((....))))))))). ( -8.00, z-score = -2.38, R) >droSim1.chr2h_random 225364 52 + 3178526 ---UACUACUACUACAGCUCGGCCAUCCUGAUAAAU-AGGAUCUCCUGGUCCUCGC- ---.............(((.((..((((((.....)-)))))..)).)))......- ( -8.90, z-score = -0.27, R) >droAna3.scaffold_13230 1660221 57 - 3602488 UUAGCAUACUGCUACAAUUCGGCCAUUCUGACCGUCAAGGAUCUCCUGAUUCUUUUU .((((.....)))).....(((.(.....).)))..(((((((....)))))))... ( -11.00, z-score = -1.56, R) >droVir3.scaffold_13324 2173639 51 - 2960039 ---UGUCGAUCUUACA-UUCGGCUAACCUGACUACA-GAUCGCCCACGAUCUUACC- ---.(((((.......-.)))))............(-(((((....))))))....- ( -11.30, z-score = -2.96, R) >droMoj3.scaffold_6117 113213 53 + 116300 ----UAUCCUGCUACAAUUCGGCCAUCCUGACUCUGAGAUAGCCCUCUAUCUCUGUU ----......(((.......)))......(((...(((((((....))))))).))) ( -8.90, z-score = -1.09, R) >consensus ____UAUACUACUACAAUUCGGCCAUCCUGACAAUU_AAGAUCCCCUGAUCUUUGC_ ..................((((.....))))......((((((....)))))).... ( -2.58 = -2.94 + 0.36)

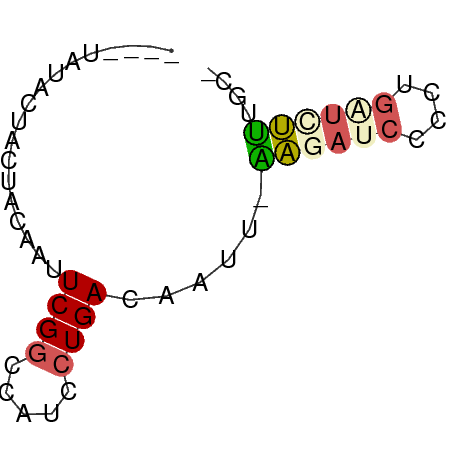
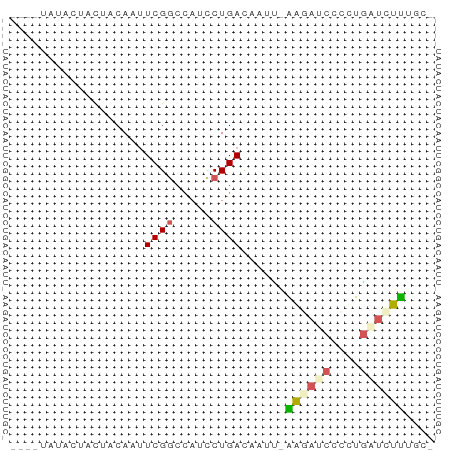
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:01:22 2011