| Sequence ID | dm3.chr2R |
|---|---|
| Location | 594,327 – 594,420 |
| Length | 93 |
| Max. P | 0.966211 |

| Location | 594,327 – 594,420 |
|---|---|
| Length | 93 |
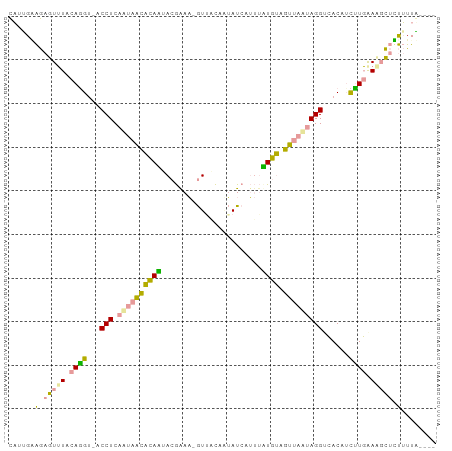
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 50.87 |
| Shannon entropy | 0.69071 |
| G+C content | 0.34899 |
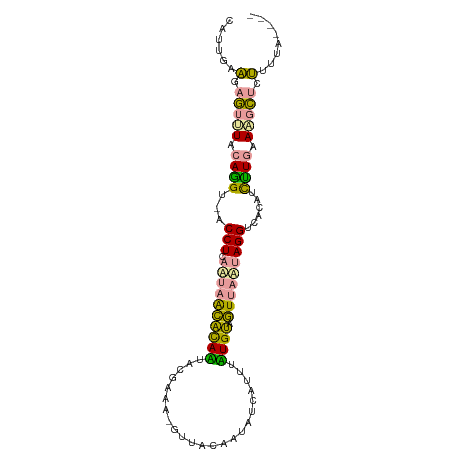
| Mean single sequence MFE | -19.50 |
| Consensus MFE | -9.12 |
| Energy contribution | -7.80 |
| Covariance contribution | -1.32 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.966211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 594327 93 + 21146708 CAUUGUAGAGUUUUCAGGUAACCUCAAUAGCACAGUACGGAA-GUAACAGUAUCAUUUCUGUAGUUAUUAGGUCACAGCCUGGACCCUGUUUUG---- ........((..(((((((.((((.((((((....(((....-)))((((........)))).))))))))))....)))))))..))......---- ( -25.40, z-score = -2.57, R) >droEre2.scaffold_4929 22107411 93 + 26641161 CAUAGAAGAGUUUACAGGUGACCUCAAUAACACAAUACGAAA-GUUCCAUUAUCAUUUUUGUAGUUAAAAGGGCACUUCUUGAAAGCUUUUUUU---- ...((((((((((.((((((.(((...((((....(((((((-((.........)))))))))))))...)))))))...)).)))))))))).---- ( -19.40, z-score = -2.05, R) >anoGam1.chr3L 30866962 95 - 41284009 UUCUGGGAAAUAUCCAAG---CCUCAGACAGCUAUUAAAAAUUAUUGUCAAAUAAACAAUGGCUUUGCUAGGUAACCUUUUUAAUGUACCGUUAUUUA .....(((....)))..(---(((.((..(((((((.....(((((....)))))..)))))))...))))))......................... ( -13.70, z-score = 0.42, R) >consensus CAUUGAAGAGUUUACAGGU_ACCUCAAUAACACAAUACGAAA_GUUACAAUAUCAUUUAUGUAGUUAAUAGGUCACAUCUUGAAAGCUCUUUUA____ ......(.(((((.((((...(((.((((((((((.......................)))).)))))))))......)))).))))).)........ ( -9.12 = -7.80 + -1.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:01:18 2011