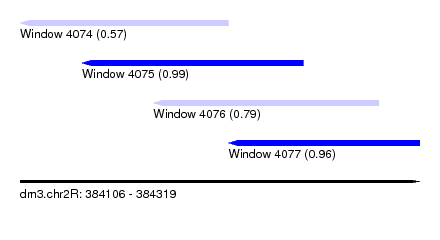
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 384,106 – 384,319 |
| Length | 213 |
| Max. P | 0.989255 |

| Location | 384,106 – 384,217 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.15 |
| Shannon entropy | 0.47261 |
| G+C content | 0.58998 |
| Mean single sequence MFE | -39.06 |
| Consensus MFE | -24.72 |
| Energy contribution | -25.16 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.568430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 384106 111 - 21146708 CGUAGCCACCACCUGGUCCUGAUCCGCACUCUGUACCACACCACAGCUACAGAGGCGACAACGAGGUGACUG--UGUUGUGGACAGCAACUCGCCGCAACACAC-UUCAGCAAC------ ....((...(((((.((..((...(((.(((((((.(........).)))))))))).)))).)))))..((--((((((((.(........))))))))))).-....))...------ ( -38.30, z-score = -2.03, R) >droAna3.scaffold_13417 814708 120 + 6960332 CGUGGCCACCACCUGGUCUUGAUCGGUGCUCUGCAGUGCUACACGACUGUAGAGGCACCUGCCGACUGACUGGCCGUUGUGGGCAGCCACUUGCCACAACGGUUCCCCAGAGCCAACAAA ..((((........((((..(...(((((((((((((........)))))))).)))))..).))))..(((((((((((((.(((....))))))))))))....)))).))))..... ( -49.10, z-score = -1.79, R) >droYak2.chrU 2863707 107 - 28119190 CGUGGCCACCACCUGGUCCCGAUCCGCACUCUGUGUGGUA-CAGAGCUACAGAAGCGACAACGAGGUGACUG--UGCUGGGGGCAGCAACGUGCCGCGGUAUGCGGUAAU---------- ((.(((((.....))))).))..((((((((((((...))-))))).(((....(((...(((((....)).--(((((....))))).)))..))).))))))))....---------- ( -37.80, z-score = 0.82, R) >droSec1.super_346 11451 112 - 13895 CGUGGACACCACCUGAUCCCGAUCCGCACUCUGCACCACAACGCAGCUACAGAGGCGACAACGAGGUGACUG--UGUUGUGAUCAUCAACGUGCCGCAACACACAUUCAGAAAC------ .((((.(((....(((((......(((.(((((................))))))))((((((((....)).--))))))))))).....))))))).................------ ( -31.99, z-score = -0.89, R) >droSim1.chr3h_random 192763 111 + 1452968 CGUGGCCACCACCUGGUCCAGAUCCGCACUCUGUACCACACCACAGCUACAGAGGCGACAACGAGGUGACUG--UGUUGUGGGCAGCAACUCGCCGCAACACAC-UUCAGCAAC------ ((((((((.....)))))......(((.(((((((.(........).))))))))))...))).(.(((.((--((((((((.(........))))))))))).-.))).)...------ ( -38.10, z-score = -0.97, R) >consensus CGUGGCCACCACCUGGUCCCGAUCCGCACUCUGUACCACAACACAGCUACAGAGGCGACAACGAGGUGACUG__UGUUGUGGGCAGCAACUUGCCGCAACACAC_UUCAGAAAC______ .(((((...((((((((....)))(((.(((((((.(........).))))))))))......)))))......(((((....)))))....)))))....................... (-24.72 = -25.16 + 0.44)

| Location | 384,139 – 384,257 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.84 |
| Shannon entropy | 0.45577 |
| G+C content | 0.55281 |
| Mean single sequence MFE | -42.07 |
| Consensus MFE | -29.53 |
| Energy contribution | -30.72 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.989255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 384139 118 - 21146708 CGGCUUAUUCCACUUGGUCACUUCAGCUAUAGUCGCUGAUCGUAGCCACCACCUGGUCCUGAUCCGCACUCUGUACCACACCACAGCUACAGAGGCGACAACGAGGUGACUG--UGUUGU .((((..........))))....((((.((((((((((((((..((((.....))))..)))))(((.(((((((.(........).)))))))))).......))))))))--))))). ( -39.90, z-score = -1.80, R) >droAna3.scaffold_13417 814748 120 + 6960332 UAGCUUACUGGGAAAGUUCAGCGGCAUCAUGAUCACUGAUCGUGGCCACCACCUGGUCUUGAUCGGUGCUCUGCAGUGCUACACGACUGUAGAGGCACCUGCCGACUGACUGGCCGUUGU ..........((..(((.((((((((....(((((..((((((((...))))..)))).)))))(((((((((((((........)))))))).)))))))))).))))))..))..... ( -51.10, z-score = -3.12, R) >droEre2.scaffold_4845 2104804 102 - 22589142 CGGCUUAUUCCAGUUGGUCACCUGAGCUAUAGUCACCGAUCGUGGCCACCACCUGGUCCCGAUCCGCACU----------------CUACAGAGGCGACAUCGAGGUGACUA--UGUUGU .((((((..((....)).....))))))((((((((((((((.(((((.....))))).)))))(((.((----------------(....)))))).......))))))))--)..... ( -40.80, z-score = -3.14, R) >droYak2.chrU 2863737 117 - 28119190 CGGCUUAUUCCAGUUGGUCACUUGAGCUAUAGUCACCGUUCGUGGCCACCACCUGGUCCCGAUCCGCACUCUGUGUGGU-ACAGAGCUACAGAAGCGACAACGAGGUGACUG--UGCUGG .((((((..((....)).....))))))(((((((((..(((.(((((.....))))).)))..(((..(((((((...-.....)).))))).))).......))))))))--)..... ( -38.90, z-score = -0.22, R) >droSec1.super_346 11485 118 - 13895 CGGCUUAUUCCAGUUGGUCACUUAAGCUAUUGUCACCGAUCGUGGACACCACCUGAUCCCGAUCCGCACUCUGCACCACAACGCAGCUACAGAGGCGACAACGAGGUGACUG--UGUUGU .((((((..((....)).....))))))...(((((((((((.(((((.....)).))))))))(((.(((((................)))))))).......))))))..--...... ( -36.69, z-score = -1.11, R) >droSim1.chr3h_random 192796 118 + 1452968 CGGCUUAUUCCAGUUGGUCACUUCAGCUAUAGUCACCGAUCGUGGCCACCACCUGGUCCAGAUCCGCACUCUGUACCACACCACAGCUACAGAGGCGACAACGAGGUGACUG--UGUUGU .((((.((....)).))))....((((.(((((((((((((.((((((.....))).)))))))(((.(((((((.(........).)))))))))).......))))))))--))))). ( -45.00, z-score = -3.02, R) >consensus CGGCUUAUUCCAGUUGGUCACUUGAGCUAUAGUCACCGAUCGUGGCCACCACCUGGUCCCGAUCCGCACUCUGUACCACAACACAGCUACAGAGGCGACAACGAGGUGACUG__UGUUGU .((((..........)))).....(((..(((((((((((((.(((((.....))))).)))))(((.(((((((.(........).)))))))))).......))))))))...))).. (-29.53 = -30.72 + 1.19)

| Location | 384,177 – 384,297 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.17 |
| Shannon entropy | 0.39428 |
| G+C content | 0.53333 |
| Mean single sequence MFE | -35.44 |
| Consensus MFE | -24.34 |
| Energy contribution | -23.42 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.790174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 384177 120 - 21146708 AACUGCCGUGAAUAUCAAGUCGGAGCUACGACCCUGCUACCGGCUUAUUCCACUUGGUCACUUCAGCUAUAGUCGCUGAUCGUAGCCACCACCUGGUCCUGAUCCGCACUCUGUACCACA ..(((..((((.....((((((((((.........))).)))))))...((....))))))..))).(((((..((.(((((..((((.....))))..))))).))...)))))..... ( -34.70, z-score = -1.95, R) >droAna3.scaffold_13417 814788 120 + 6960332 AACCGCCGCGGCAGUAUAGCUGAAGCUAAUACCCUGCUAUUAGCUUACUGGGAAAGUUCAGCGGCAUCAUGAUCACUGAUCGUGGCCACCACCUGGUCUUGAUCGGUGCUCUGCAGUGCU ....(((((..(((((.(((((((((.........))).))))))))))).((....)).))))).....((.(((((((((.(((((.....))))).))))))))).))......... ( -45.50, z-score = -2.26, R) >droYak2.chrU 2863774 120 - 28119190 AACCACUGUCAAUAUCAAGUCAGAGCUACGACCCUGCUACCGGCUUAUUCCAGUUGGUCACUUGAGCUAUAGUCACCGUUCGUGGCCACCACCUGGUCCCGAUCCGCACUCUGUGUGGUA .(((((........((((((...(((.........)))(((((((......))))))).))))))...((((....((.(((.(((((.....))))).)))..))....))))))))). ( -32.20, z-score = -0.32, R) >droSec1.super_346 11523 120 - 13895 ACCUAACGUGAAUAUCAAGUCGGAGCUACGACCCUGCUACCGGCUUAUUCCAGUUGGUCACUUAAGCUAUUGUCACCGAUCGUGGACACCACCUGAUCCCGAUCCGCACUCUGCACCACA ..(((((..(((((...(((((((((.........))).)))))))))))..)))))........((...(((....(((((.(((((.....)).)))))))).)))....))...... ( -30.60, z-score = -1.44, R) >droSim1.chr3h_random 192834 120 + 1452968 AACUGCCGUGAAUAUCAAGUCGGAGCUACGACCCUGCUACCGGCUUAUUCCAGUUGGUCACUUCAGCUAUAGUCACCGAUCGUGGCCACCACCUGGUCCAGAUCCGCACUCUGUACCACA ....(((((((.....((((((((((.........))).))))))).......(((((.(((........))).)))))))))))).......((((.((((.......)))).)))).. ( -34.20, z-score = -1.23, R) >consensus AACUGCCGUGAAUAUCAAGUCGGAGCUACGACCCUGCUACCGGCUUAUUCCAGUUGGUCACUUCAGCUAUAGUCACCGAUCGUGGCCACCACCUGGUCCCGAUCCGCACUCUGUACCACA .......(((......((((((((((.........))).))))))).........(((.(((........))).)))(((((.(((((.....))))).)))))............))). (-24.34 = -23.42 + -0.92)

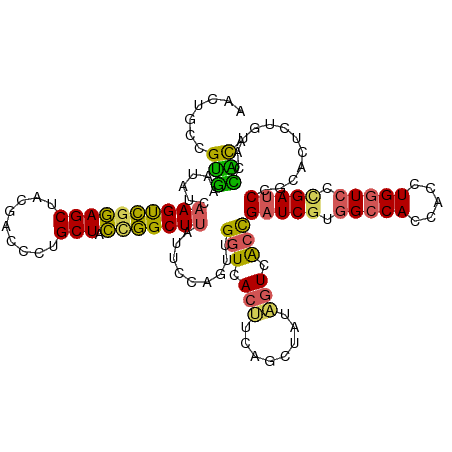
| Location | 384,217 – 384,319 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.84 |
| Shannon entropy | 0.56053 |
| G+C content | 0.44265 |
| Mean single sequence MFE | -28.26 |
| Consensus MFE | -14.60 |
| Energy contribution | -14.32 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.962229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 384217 102 - 21146708 ----------------UAAAGUGACGUUAUCGUAAA-AC-AACUGCCGUGAAUAUCAAGUCGGAGCUACGACCCUGCUACCGGCUUAUUCCACUUGGUCACUUCAGCUAUAGUCGCUGAU ----------------...(((((((((.......)-))-.......(((((((...(((((((((.........))).)))))))))).)))...))))))(((((.......))))). ( -28.50, z-score = -2.21, R) >droAna3.scaffold_13417 814828 120 + 6960332 UAAAGGCACCUUAUCUGAGACUGGUGUUUUCAUCAUUGUCAACCGCCGCGGCAGUAUAGCUGAAGCUAAUACCCUGCUAUUAGCUUACUGGGAAAGUUCAGCGGCAUCAUGAUCACUGAU ..((((((((............)))))))).((((.(((((...(((((..(((((.(((((((((.........))).))))))))))).((....)).)))))....))).)).)))) ( -35.60, z-score = -1.43, R) >droYak2.chrU 2863814 119 - 28119190 UAAAGGCCCAUCAUCGUAAUCGAAUCUUUUAUUUAAUAU-AACCACUGUCAAUAUCAAGUCAGAGCUACGACCCUGCUACCGGCUUAUUCCAGUUGGUCACUUGAGCUAUAGUCACCGUU ....((.......(((....)))................-....(((((.....((((((...(((.........)))(((((((......))))))).))))))...)))))..))... ( -19.00, z-score = 0.18, R) >droSec1.super_346 11563 102 - 13895 ----------------UAAGGUGACGUUAUCAUAAA-AU-ACCUAACGUGAAUAUCAAGUCGGAGCUACGACCCUGCUACCGGCUUAUUCCAGUUGGUCACUUAAGCUAUUGUCACCGAU ----------------...(((((((.((.(.(((.-..-(((.(((..(((((...(((((((((.........))).)))))))))))..))))))...))).).)).)))))))... ( -30.30, z-score = -3.12, R) >droSim1.chr3h_random 192874 102 + 1452968 ----------------UAAAGUGACGUUAUCGUUAA-AU-AACUGCCGUGAAUAUCAAGUCGGAGCUACGACCCUGCUACCGGCUUAUUCCAGUUGGUCACUUCAGCUAUAGUCACCGAU ----------------....((((((((((......-))-)))....(.(((((...(((((((((.........))).))))))))))))((((((.....))))))...))))).... ( -27.90, z-score = -2.05, R) >consensus ________________UAAAGUGACGUUAUCAUAAA_AU_AACUGCCGUGAAUAUCAAGUCGGAGCUACGACCCUGCUACCGGCUUAUUCCAGUUGGUCACUUCAGCUAUAGUCACCGAU ...................((((((...............................((((((((((.........))).)))))))...((....))..............))))))... (-14.60 = -14.32 + -0.28)

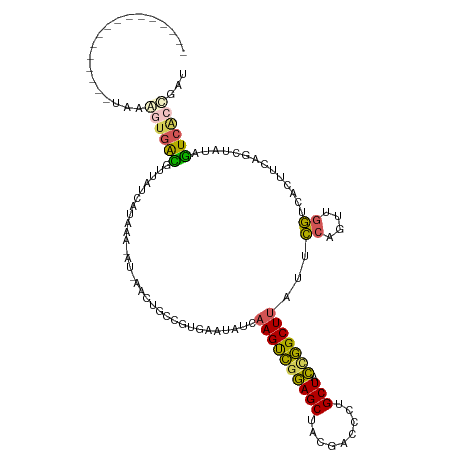
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:01:13 2011