
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 378,561 – 378,648 |
| Length | 87 |
| Max. P | 0.920328 |

| Location | 378,561 – 378,648 |
|---|---|
| Length | 87 |
| Sequences | 5 |
| Columns | 89 |
| Reading direction | forward |
| Mean pairwise identity | 64.52 |
| Shannon entropy | 0.64673 |
| G+C content | 0.52984 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -9.52 |
| Energy contribution | -9.56 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
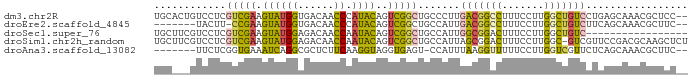
>dm3.chr2R 378561 87 + 21146708 UGCACUGUCCUCGUCGAAGUAUGGUGACAACCCAUACAGUCGGCUGCCCUUGACGGCCUUUCCUUGGCUGUCCUGAGCAAACGCUCC-- .((.........(((((.((((((.......))))))..)))))(((.(..(((((((.......)))))))..).)))...))...-- ( -31.00, z-score = -2.80, R) >droEre2.scaffold_4845 984210 79 + 22589142 -------UACUU-CCGAAGUAUGGUGACAACCCAUACAGUCGGCUGCCAUUGACGGCCUUUCCUUGGCUGUCUUCAGCAAACGCUUC-- -------.....-((((.((((((.......))))))..)))).(((....(((((((.......)))))))....)))........-- ( -26.50, z-score = -3.12, R) >droSec1.super_76 64049 72 - 142355 UGCUUCGUCCUCGUCGAAGUAUGGAGACAACCAAUACAGUCGGCUGCCAUUGGCGGACUUUCCUUGGCUGUC----------------- (((((((.......)))))))....((((.((((...((((.(((......))).))))....)))).))))----------------- ( -23.90, z-score = -1.68, R) >droSim1.chr2h_random 1176787 88 - 3178526 UGCUUCGUCCUCGUCGAAGUAUGGAGACAACCAAUACAGUCGGCUGCCAUUAGCGGACUUUCCUUGGC-GUCGUUCCGACGCAAGCUCU (((((((.......))))))).((((.(...(((...((((.((((....)))).))))....)))((-((((...))))))..))))) ( -28.70, z-score = -1.52, R) >droAna3.scaffold_13082 1439562 79 + 3919855 -------UUCUCGGUGAAAUCAGGCGCUCUUCAAGGUAGGUGAGU-CCAUUUAAGGUUUUUCCUUGGUCGUUCUCAGCAAACGCUUC-- -------..............(((((((......))).(.((((.-.(..((((((.....))))))..)..)))).)....)))).-- ( -15.90, z-score = 0.05, R) >consensus UGC___GUCCUCGUCGAAGUAUGGAGACAACCAAUACAGUCGGCUGCCAUUGACGGACUUUCCUUGGCUGUCCUCAGCAAACGCUCC__ ............(((((.((((((......)).))))..))))).......(((((((.......)))))))................. ( -9.52 = -9.56 + 0.04)

| Location | 378,561 – 378,648 |
|---|---|
| Length | 87 |
| Sequences | 5 |
| Columns | 89 |
| Reading direction | reverse |
| Mean pairwise identity | 64.52 |
| Shannon entropy | 0.64673 |
| G+C content | 0.52984 |
| Mean single sequence MFE | -24.64 |
| Consensus MFE | -9.18 |
| Energy contribution | -11.22 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.511430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
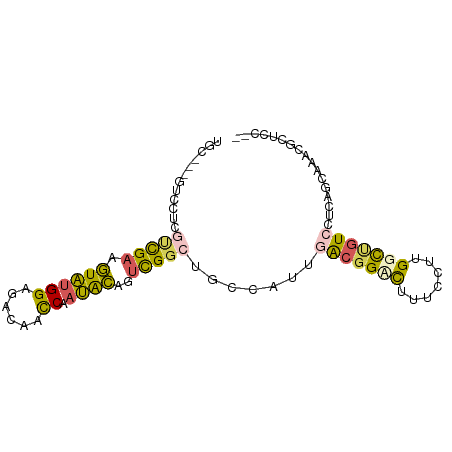
>dm3.chr2R 378561 87 - 21146708 --GGAGCGUUUGCUCAGGACAGCCAAGGAAAGGCCGUCAAGGGCAGCCGACUGUAUGGGUUGUCACCAUACUUCGACGAGGACAGUGCA --...(((.((((((..(((.(((.......))).)))..)))))).(((..((((((.......)))))).)))..........))). ( -29.10, z-score = -0.79, R) >droEre2.scaffold_4845 984210 79 - 22589142 --GAAGCGUUUGCUGAAGACAGCCAAGGAAAGGCCGUCAAUGGCAGCCGACUGUAUGGGUUGUCACCAUACUUCGG-AAGUA------- --...((..((((((..(((.(((.......))).)))..))))))((((..((((((.......)))))).))))-..)).------- ( -26.70, z-score = -1.85, R) >droSec1.super_76 64049 72 + 142355 -----------------GACAGCCAAGGAAAGUCCGCCAAUGGCAGCCGACUGUAUUGGUUGUCUCCAUACUUCGACGAGGACGAAGCA -----------------(((((((((....((((.((........)).))))...)))))))))......(((((.......))))).. ( -24.40, z-score = -2.20, R) >droSim1.chr2h_random 1176787 88 + 3178526 AGAGCUUGCGUCGGAACGAC-GCCAAGGAAAGUCCGCUAAUGGCAGCCGACUGUAUUGGUUGUCUCCAUACUUCGACGAGGACGAAGCA ...((((.(((((((..(((-(((((....((((.(((......))).))))...)))).)))))))...(((....))))))))))). ( -30.90, z-score = -1.61, R) >droAna3.scaffold_13082 1439562 79 - 3919855 --GAAGCGUUUGCUGAGAACGACCAAGGAAAAACCUUAAAUGG-ACUCACCUACCUUGAAGAGCGCCUGAUUUCACCGAGAA------- --...((((((..((((..((...((((.....))))...)).-.)))).(......)..))))))................------- ( -12.10, z-score = 0.12, R) >consensus __GAAGCGUUUGCUGAGGACAGCCAAGGAAAGGCCGUCAAUGGCAGCCGACUGUAUUGGUUGUCACCAUACUUCGACGAGGAC___GCA .................(((((((((((.....)).......((((....)))).)))))))))......................... ( -9.18 = -11.22 + 2.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:01:09 2011