
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 300,437 – 300,514 |
| Length | 77 |
| Max. P | 0.770099 |

| Location | 300,437 – 300,514 |
|---|---|
| Length | 77 |
| Sequences | 9 |
| Columns | 86 |
| Reading direction | forward |
| Mean pairwise identity | 57.09 |
| Shannon entropy | 0.88729 |
| G+C content | 0.41699 |
| Mean single sequence MFE | -15.52 |
| Consensus MFE | -6.04 |
| Energy contribution | -6.14 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 2.08 |
| Mean z-score | -0.71 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.770099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
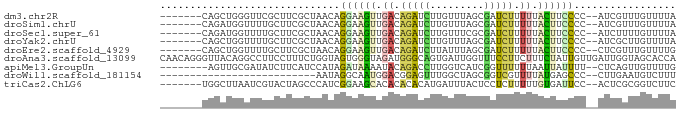
>dm3.chr2R 300437 77 + 21146708 -------CAGCUGGGUUCGCUUCGCUAACAGGAAGUUGACAGAUCUUGUUUAGCGAUCUUUUUACUUCCCC--AUCGUUUGUUUUA -------..(.((((((.((...)).))).((((((.((.(((((.........))))).)).))))))))--).).......... ( -17.40, z-score = -1.09, R) >droSim1.chrU 7156434 77 + 15797150 -------CAGAUGGUUUUGCUUCGCUAACAGGAAGUUGACAGAUCUUGUUUAGCGAUCUUUUUACUUCCCC--AUCGUUUGUUUUA -------..(((((....((...)).....((((((.((.(((((.........))))).)).))))))))--))).......... ( -18.80, z-score = -2.35, R) >droSec1.super_61 122850 77 + 189864 -------CAGAUGGUUUUGCUUCGCUAACAGGAAGUUGACAGAUCUUGUUUCGCGAUCUUUUUACUUCCCC--AUCUUUUGUUUUA -------.((((((....((...)).....((((((.((.(((((.........))))).)).))))))))--))))......... ( -18.90, z-score = -2.65, R) >droYak2.chrU 10224659 77 + 28119190 -------CAGCUGGUUUUGCUUCGCUAACAGGAAGUUGACAGAUCUUGUUUAGCGAUCUUUUUACUUCCCC--AUCGCUUGUUUUA -------.(((.......)))..((.....((((((.((.(((((.........))))).)).))))))..--...))........ ( -15.70, z-score = -0.68, R) >droEre2.scaffold_4929 1838311 77 - 26641161 -------CAGCUGGUUUUGCUUCGCUAACAGGAAGUUGACAGAUCUUAUUUAGCGAUCUUUUUACUUCCCC--CUCGUUUGUUUUG -------.(((.((.....))..)))....((((((.((.(((((.........))))).)).))))))..--............. ( -15.00, z-score = -0.96, R) >droAna3.scaffold_13099 994553 86 + 3324263 CAACAGGGUUACAGGCCUUCCUUUCUGGUAGUGGGUAGAUGGGCAGUGAUUGGUUUCCUUCUUUCUAUUGUUGAUUGGUAGCACCA .....(((((((..((((.((.....))....)))).(((.((((((((..((......))..)).)))))).))).))))).)). ( -22.20, z-score = 0.33, R) >apiMel3.GroupUn 316374847 76 - 399230636 --------AGUUGCGAUAUCUUCAUCCAUAGAUAAAAUACAGACCUUGGUCAUCGGUUUUUUAAUUAUUUU--CUCAGUUGUUUUG --------.....((((((((........))))).......(((....))).)))................--............. ( -8.60, z-score = 0.20, R) >droWil1.scaffold_181154 1714036 58 + 4610121 --------------------------AAUAGGCAAUGGACGGAGUUUGGCUAGCGGUCGUUUUAUGAGCCC--CUUGAAUGUCUUU --------------------------...(((((.(.((.((.(((((...(((....)))...)))))))--.)).).))))).. ( -11.70, z-score = 0.58, R) >triCas2.ChLG6 13062924 77 + 13544221 -------UGGCUUAAUCGUACUAGCCCAUCGGAAGCACACACACAUGAUUUACUCCUCUUUUUGUGAUUCC--ACUCGCGGUCUUC -------.((((..........))))....((((.((((.......((.....)).......)))).))))--............. ( -11.34, z-score = 0.20, R) >consensus _______CAGCUGGUUUUGCUUCGCUAACAGGAAGUUGACAGAUCUUGUUUAGCGAUCUUUUUACUUCCCC__AUCGUUUGUUUUA ..............................((((((.((.(((((.........))))).)).))))))................. ( -6.04 = -6.14 + 0.10)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:01:01 2011