
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,129,131 – 2,129,270 |
| Length | 139 |
| Max. P | 0.772134 |

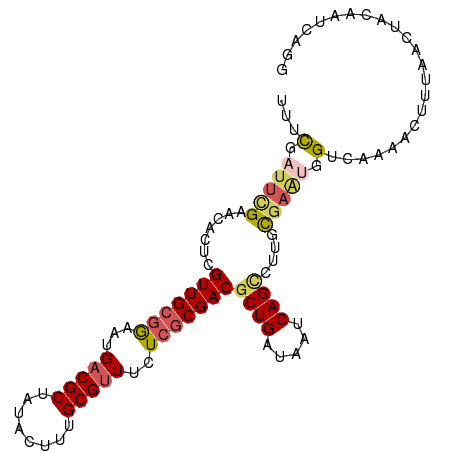
| Location | 2,129,131 – 2,129,232 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 85.94 |
| Shannon entropy | 0.25077 |
| G+C content | 0.41980 |
| Mean single sequence MFE | -25.24 |
| Consensus MFE | -18.68 |
| Energy contribution | -19.44 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.772134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2129131 101 - 23011544 UUUUGAUUCGAACACUCGUUGCGGAAUGUCGCUAUACUUUGCGUUUCUCACGACACUGAUAAUCAGCCUUGUGAAUAGUCAAUACUUUAACUACAAUCAGG ..((((((.(..(((..((((.((((...(((........))).))).).)))).(((.....)))....)))..((((.((....)).))))))))))). ( -17.90, z-score = -0.14, R) >droEre2.scaffold_4929 2163423 101 - 26641161 UUUCAUUUUGAACACUCGUUGCGAAAUGACGCUAUACCUUGCGUUUCUGGCGACGCUGAUAAUCAGUUUUGCGAAUUGCAAACACAAUAACAAAAAUCAGG ..(((((((((((....))).))))))))(((........)))...(((((((.((((.....)))).))))..((((......)))).........))). ( -23.20, z-score = -1.49, R) >droYak2.chr2L 2100541 101 - 22324452 UUUUAUUCGGAACACUCGUUGCGAAAUGACGCUUUACUUUGCGUUUCUCGCGACGCUGAUAAUCAGCUUUGCACUUUGUCAAAACUUAAACAACAAACAGG .........((.((...(((((((...(((((........)))))..)))))))((((.....)))).........))))..................... ( -23.60, z-score = -2.09, R) >droSec1.super_14 2058661 101 - 2068291 UUUCGAUUCGAAUACUCGUUGCGGAAUGACGCUAUACUUUGCGUUUCUCGCGACGCUGAUAAUCAGCCUUGCGAGUGGUCAAAACUUUAACUACAAUCAGG .........((.((((((((((((...(((((........)))))..)))))))((((.....)))).....))))).))..................... ( -29.90, z-score = -2.59, R) >droSim1.chr2L 2082577 101 - 22036055 UUUCGAUUCGAACACUCGUUGCGGAAUGACGCUAUACUUUGCGUUUCUCGCGACGCUGAUAAUCAGCCUUGCGAGUGGUCAAAACUUUAACUACAAUCAGG .........((.((((((((((((...(((((........)))))..)))))))((((.....)))).....))))).))..................... ( -31.60, z-score = -3.12, R) >consensus UUUCGAUUCGAACACUCGUUGCGGAAUGACGCUAUACUUUGCGUUUCUCGCGACGCUGAUAAUCAGCCUUGCGAAUGGUCAAAACUUUAACUACAAUCAGG ...(.(((((.......(((((((...(((((........)))))..)))))))((((.....))))....))))).)....................... (-18.68 = -19.44 + 0.76)


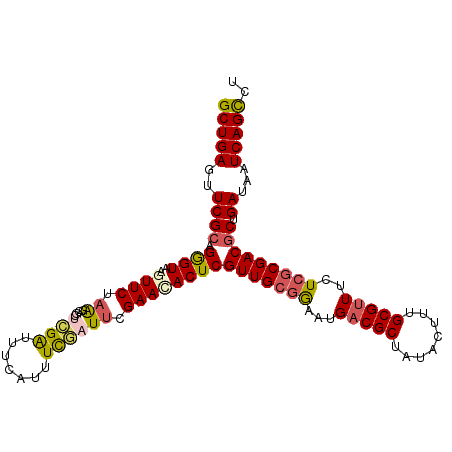
| Location | 2,129,163 – 2,129,270 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 88.88 |
| Shannon entropy | 0.19642 |
| G+C content | 0.45794 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -26.76 |
| Energy contribution | -27.24 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.634384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2129163 107 - 23011544 GCUGAGUUCGCAGGGUAAGUUCUAAGUACUCGAUUUCAUUUUGAUUCGAACACUCGUUGCGGAAUGUCGCUAUACUUUGCGUUUCUCACGACACUGAUAAUCAGCCU (((((..((((((((((.......(((..((((..((.....)).))))..)))....((((....))))..))))))))(((......)))...))...))))).. ( -26.70, z-score = -0.81, R) >droEre2.scaffold_4929 2163455 107 - 26641161 GCUGAGUUCGCGGAGUAAAUUCUAGGCGCUUGAUUUCAUUUCAUUUUGAACACUCGUUGCGAAAUGACGCUAUACCUUGCGUUUCUGGCGACGCUGAUAAUCAGUUU ((((((((((((.((......))...))).(((.......)))....))))...((((((.(...(((((........)))))..).)))))).......))))).. ( -27.30, z-score = -0.38, R) >droYak2.chr2L 2100573 107 - 22324452 GCUGAGUUCGCAGAGUAAAUUCUAAGAGCUCGGAUUCAUUUUAUUCGGAACACUCGUUGCGAAAUGACGCUUUACUUUGCGUUUCUCGCGACGCUGAUAAUCAGCUU .((((((((..((((....))))..))))))))......................(((((((...(((((........)))))..)))))))((((.....)))).. ( -37.00, z-score = -3.10, R) >droSec1.super_14 2058693 107 - 2068291 GCUGAGUUCGCAGGGUAAGUUCUAAGCGCUCGAUUUCAUUUCGAUUCGAAUACUCGUUGCGGAAUGACGCUAUACUUUGCGUUUCUCGCGACGCUGAUAAUCAGCCU (((((..((((.(((((.((.....))..((((..((.....)).)))).)))))(((((((...(((((........)))))..))))))))).))...))))).. ( -35.20, z-score = -1.82, R) >droSim1.chr2L 2082609 107 - 22036055 GCUGAGUUCGCCGGGUAAGUUCUAAGCGCUCGAUUUCAUUUCGAUUCGAACACUCGUUGCGGAAUGACGCUAUACUUUGCGUUUCUCGCGACGCUGAUAAUCAGCCU (((((((((((..((......))..))..((((.......))))...))))...((((((((...(((((........)))))..)))))))).......))))).. ( -34.70, z-score = -1.57, R) >consensus GCUGAGUUCGCAGGGUAAGUUCUAAGCGCUCGAUUUCAUUUCGAUUCGAACACUCGUUGCGGAAUGACGCUAUACUUUGCGUUUCUCGCGACGCUGAUAAUCAGCCU (((((..((((.((((..((((.((....((((.......)))))).))))))))(((((((...(((((........)))))..))))))))).))...))))).. (-26.76 = -27.24 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:10:17 2011