
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 230,660 – 230,840 |
| Length | 180 |
| Max. P | 0.899603 |

| Location | 230,660 – 230,772 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.72 |
| Shannon entropy | 0.60210 |
| G+C content | 0.56734 |
| Mean single sequence MFE | -42.64 |
| Consensus MFE | -16.82 |
| Energy contribution | -16.34 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.899603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
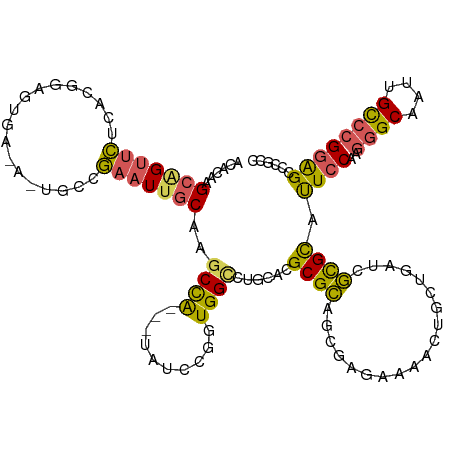
>dm3.chr2R 230660 112 + 21146708 ACCCAAGCAGUUCUCACUGAGUAAAAGUGAAGAAUUGCAAGCCA---UAUCCCGUGGCCUGCACGCGCGGUCAGGAAAAUGCUGGUUGUGCACUCCAGGGGCAACGGCCAGGAGC----- ..((..((((((((((((.......)))).))))))))..(((.---...(((.(((..(((((((.(((.((......)))))).))))))..))).)))....)))..))...----- ( -44.40, z-score = -2.26, R) >droSec1.super_79 15328 117 + 133581 ACACAAGCAGUUCUCACGGAGUCAGACUGCCGAAUUGCAAGCUG---UAUCCGGUGGCCUGCACGCGCAGCGAGGAUAUUGCUGAUCGCGCAUUCCAAGGGCAAUUGCCCGGAGUCCGCC ......((((..(.(.((((..(((..(((......)))..)))---..))))).)..)))).....((((((.....))))))...(((.(((((..((((....))))))))).))). ( -45.70, z-score = -1.60, R) >droSim1.chrU 7081155 107 + 15797150 ACACAAGCAGUUCUCAUGGAGU---------GAAUC-CAGGCCA---UAUCCAGUGGUCUGCACGCGCACUAAGCAAACUGCUGAUCGCGUAUUCCAGGUGCAAUUGCCCGGAGCCCGCA .....(((((((..(.(((.((---------(....-(((((((---(.....))))))))....))).))).)..)))))))....(((..((((.((.((....))))))))..))). ( -40.50, z-score = -2.55, R) >droEre2.scaffold_4784 24528627 120 + 25762168 GCACAAGCGGUUCUUACGGAAAGAUACCGCUGAAUUGCAGGCCACCAUAUCGGAUGGCUUACACGCGCAUCGGAAGAACUGCUGAUCGCGCAUUCCAAGGGCAAUUGCCCGGAGCCCGGU (((..((((((((((.....)))).))))))....)))((((((((.....)).))))))....(((((((((........))))).)))).((((..((((....))))))))...... ( -46.50, z-score = -2.44, R) >droAna3.scaffold_13230 550919 113 - 3602488 GCGAAUGCAGAAUUUAGGAAAAG----UGCCAAAUUGCACACCG---GAUCCGGUGGCCCGCACGCGUAACGAAAGAGGUGCUGACUACGCAUUCCAAGGGCAACUGUCCGCGGACCAGC ((...(((........((((..(----(((......((.(((((---....)))))))..))))((((..(....).(((....))))))).))))..((((....))))))).....)) ( -36.10, z-score = -0.57, R) >consensus ACACAAGCAGUUCUCACGGAGUGA_A_UGCCGAAUUGCAAGCCA___UAUCCGGUGGCCUGCACGCGCAGCGAGAAAACUGCUGAUCGCGCAUUCCAAGGGCAAUUGCCCGGAGCCCGCC ......(((((((..................)))))))..((((..........))))......((((...................)))).((((..((((....))))))))...... (-16.82 = -16.34 + -0.48)

| Location | 230,737 – 230,840 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.34 |
| Shannon entropy | 0.55997 |
| G+C content | 0.61911 |
| Mean single sequence MFE | -48.26 |
| Consensus MFE | -14.57 |
| Energy contribution | -17.32 |
| Covariance contribution | 2.75 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.742791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
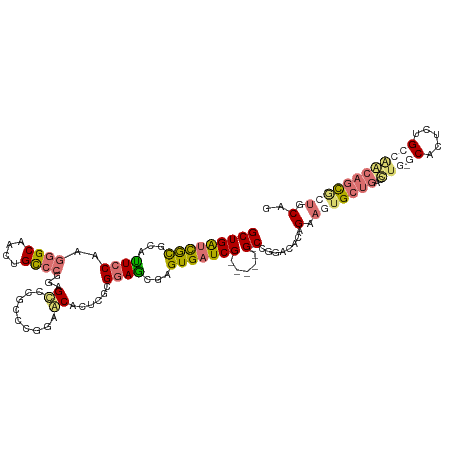
>dm3.chr2R 230737 103 + 21146708 GCUGGUUGUGCACUCCAGGGGCAACGGCCAGGAGC--------AGCA---GCGGAGCGAGUGAUCG-----GACCGGACGAAGAACUGCUGACCUG-GCUCUUUGGCGGCAGCACUACAG .(((...((((.(.((((((.....(((((((..(--------((((---((((..(((....)))-----..))).........)))))).))))-))))))))).)...))))..))) ( -42.20, z-score = -0.98, R) >droSec1.super_79 15405 114 + 133581 GCUGAUCGCGCAUUCCAAGGGCAAUUGCCCGGAGUCCGCCCGAAACACUCGCGGAGCGAGUGAUCG-----G-CCGGAUACAGGAGUGCUGACUUAAGCGCUCUGCCGAAAACGCUGCAA ((((((((((.(((((..((((....))))))))).))).......((((((...)))))))))))-----)-)(((.....((((((((......)))))))).)))............ ( -51.50, z-score = -3.26, R) >droSim1.chrU 7081222 113 + 15797150 GCUGAUCGCGUAUUCCAGGUGCAAUUGCCCGGAGCCCGCACGGAGCACUCGCGGAGCGAGUGAUCG-----G-CCUGACGCAGAAGUGCUGACCUG-GCCUUCUGCCAGCAGUUCUGCUG ((((((((((..((((.((.((....))))))))..))).......((((((...)))))))))))-----)-).....(((((((.(((.....)-)))))))))(((((....))))) ( -52.30, z-score = -2.40, R) >droAna3.scaffold_13230 550992 118 - 3602488 GCUGACUACGCAUUCCAAGGGCAACUGUCCGCGGACCAGCCCGGACACUUGCGCAACAAGUGAUCGUUGCUGGCCGGAUACGGAA-UGCUGAAACU-GCAACUGGGUAACUGCGCCGCAG ((.......(((((((..((((....)))).(((.(((((.(((.((((((.....)))))).)))..)))))))).....))))-))).......-))..((((((......))).))) ( -47.04, z-score = -2.55, R) >consensus GCUGAUCGCGCAUUCCAAGGGCAACUGCCCGGAGCCCGCCCGGAACACUCGCGGAGCGAGUGAUCG_____G_CCGGACACAGAAGUGCUGACCUG_GCACUCUGCCAACAGCGCUGCAG ((((.........(((..((((....))))...............((((((.....)))))).............)))....(((((((........)))))))..)))).......... (-14.57 = -17.32 + 2.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:01:00 2011