| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 3,213,788 – 3,213,846 |
| Length | 58 |
| Max. P | 0.581315 |

| Location | 3,213,788 – 3,213,846 |
|---|---|
| Length | 58 |
| Sequences | 4 |
| Columns | 61 |
| Reading direction | forward |
| Mean pairwise identity | 62.33 |
| Shannon entropy | 0.60190 |
| G+C content | 0.34678 |
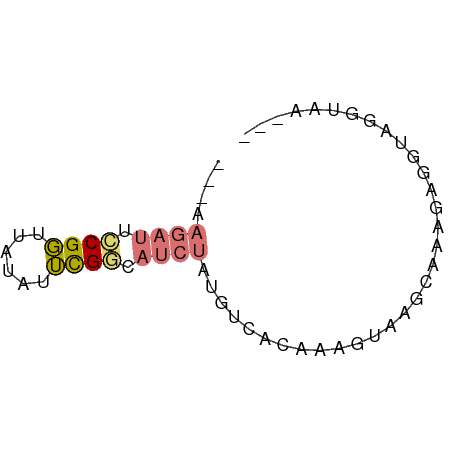
| Mean single sequence MFE | -8.99 |
| Consensus MFE | -4.04 |
| Energy contribution | -4.47 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.581315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
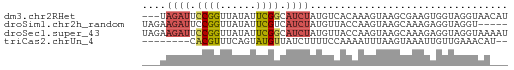
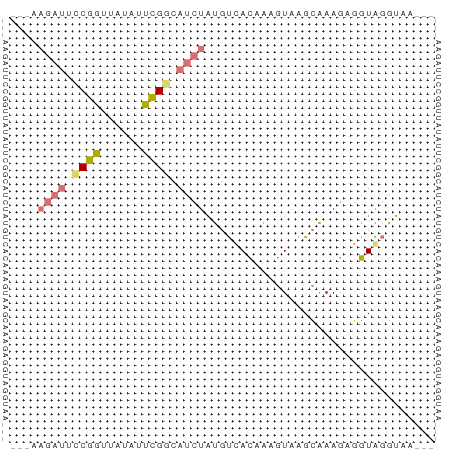
>dm3.chr2RHet 3213788 58 + 3288761 ---UAGAUUCCGGUUAUAUUCGGCAUCUAUGUCACAAAGUAAGCGAAGUGGUAGGUAACAU ---(((((.((((......)))).)))))(..(((...(....)...)))..)........ ( -11.60, z-score = -1.46, R) >droSim1.chr2h_random 845424 56 + 3178526 UAGAAGAUUCCGGUUAUAUUCGUCAUCUAUGUUACCAAGUAAGCAAAGAGGUAGGU----- ....((((..(((......)))..))))...(((((.............)))))..----- ( -7.42, z-score = -0.28, R) >droSec1.super_43 190561 61 + 327307 UAGAAGAUUCCGGUUAUAUUCGGCAUCUAUGUUACCAAGUAAGCAAAGAGGUAGGUAAAAU ....((((.((((......)))).))))...(((((.................)))))... ( -10.53, z-score = -1.29, R) >triCas2.chrUn_4 259093 51 + 1175385 --------CACGUUUCAGUAUGUUAUCUUUUCCAAAAUUUAAGUAAAUUGUUGAAACAU-- --------...(((((((((..(((.(((...........))))))..)))))))))..-- ( -6.40, z-score = -1.34, R) >consensus ___AAGAUUCCGGUUAUAUUCGGCAUCUAUGUCACAAAGUAAGCAAAGAGGUAGGUAA___ ....((((.((((......)))).))))................................. ( -4.04 = -4.47 + 0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:00:54 2011