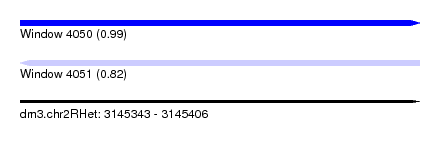
| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 3,145,343 – 3,145,406 |
| Length | 63 |
| Max. P | 0.993184 |

| Location | 3,145,343 – 3,145,406 |
|---|---|
| Length | 63 |
| Sequences | 10 |
| Columns | 69 |
| Reading direction | forward |
| Mean pairwise identity | 61.81 |
| Shannon entropy | 0.79699 |
| G+C content | 0.38799 |
| Mean single sequence MFE | -11.87 |
| Consensus MFE | -5.85 |
| Energy contribution | -4.81 |
| Covariance contribution | -1.04 |
| Combinations/Pair | 2.29 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.993184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
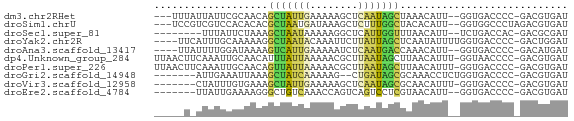
>dm3.chr2RHet 3145343 63 + 3288761 ---UUUAUUAUUCGCAACAGCUAUUGAAAAAGCUCAAUAGCUAAACAUU--GGUGACCCC-GACGUGAU ---........((((...(((((((((......))))))))).....((--((.....))-)).)))). ( -16.70, z-score = -3.75, R) >droSim1.chrU 6574800 64 + 15797150 ---UCCGUCGUCCACACACGCUAAUGAUAAAGCUCUUUGGCUACACAUU--GGUGGCCCUAGACGUGAU ---((((((..........(((........))).....((((((.....--.))))))...)))).)). ( -17.00, z-score = -1.41, R) >droSec1.super_81 106974 58 + 123496 --------UUUAUUCUAAAGCUAAUAAAAAGGCUCAUUGGUUUAACAUU--UCUGACCAC-GACGCGAU --------.......................((((..(((((.......--...))))).-)).))... ( -6.00, z-score = 0.07, R) >droYak2.chr2R 266274 64 + 21139217 ----UUCAUUUGCAAAAAGGCUAAUACAAAUUCUUAUUAGCUCAAUAUUUUGGUGACCCC-GACUGGAU ----.(((((........((((((((........)))))))).........)))))((..-....)).. ( -9.43, z-score = -0.34, R) >droAna3.scaffold_13417 6814960 62 + 6960332 ----UUAUUUUGGAUAAAAGUCAUUGAAAAAUCUCAAUGACCAAACAUU--GGUGACCCC-GACAUGAU ----((((.((((......((((((((......))))))))........--(....).))-)).)))). ( -14.50, z-score = -2.36, R) >dp4.Unknown_group_284 5815 67 - 17202 UUAACUUCAAAUUGCAACAUUUAUUAAAAACGCUUAAUAGCUUAACAUUU-GGUAACCCC-GACGUGAU ..........((..(................(((....))).......((-((.....))-)).)..)) ( -6.00, z-score = -0.06, R) >droPer1.super_226 23616 67 + 38019 UUAACUUCAAAUUGCAACAGUUAUUAAAAACGCUUAAUAGCUUAACAUUU-GGUGACCCC-GACGUGAU ..........((..(...(((((((((......)))))))))......((-((.....))-)).)..)) ( -9.50, z-score = -0.89, R) >droGri2.scaffold_14948 4364 59 + 44767 -------AUUGAAAUUAAAGCUAUCAAAAAG--CUGAUAGCGCAAACCUCUGGUGACCCC-GACGUGAU -------............(((((((.....--.)))))))....((.((.((....)).-)).))... ( -11.70, z-score = -2.05, R) >droVir3.scaffold_12958 3369160 60 - 3547706 -------CUAUUUGUGAAAGCUAUUGAAAAAGCUCAAUAGCGCAACAUUU-GGUGACCCC-GACGUGAU -------(((..(((....((((((((......))))))))...)))..)-)).......-........ ( -14.60, z-score = -2.48, R) >droEre2.scaffold_4784 25734147 59 + 25762168 -------UUAUUGAAAAGGGCUGUCAAACCAGUCAGUCCUCGUAACAUU--GGUGACCCC-GACGUGAU -------.........(((((((.(......).)))))))....((.((--((.....))-)).))... ( -13.30, z-score = -0.56, R) >consensus _____U_UUAUUUACAAAAGCUAUUAAAAAAGCUCAAUAGCUUAACAUU__GGUGACCCC_GACGUGAU ...................(((((((........)))))))............................ ( -5.85 = -4.81 + -1.04)

| Location | 3,145,343 – 3,145,406 |
|---|---|
| Length | 63 |
| Sequences | 10 |
| Columns | 69 |
| Reading direction | reverse |
| Mean pairwise identity | 61.81 |
| Shannon entropy | 0.79699 |
| G+C content | 0.38799 |
| Mean single sequence MFE | -11.69 |
| Consensus MFE | -5.77 |
| Energy contribution | -5.58 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.87 |
| Mean z-score | -0.47 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.822524 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2RHet 3145343 63 - 3288761 AUCACGUC-GGGGUCACC--AAUGUUUAGCUAUUGAGCUUUUUCAAUAGCUGUUGCGAAUAAUAAA--- .((((((.-((.....))--.)))).(((((((((((....)))))))))))....))........--- ( -16.20, z-score = -2.29, R) >droSim1.chrU 6574800 64 - 15797150 AUCACGUCUAGGGCCACC--AAUGUGUAGCCAAAGAGCUUUAUCAUUAGCGUGUGUGGACGACGGA--- ....((((((.((((((.--...)))..))).....(((........))).....)))))).....--- ( -15.30, z-score = 0.15, R) >droSec1.super_81 106974 58 - 123496 AUCGCGUC-GUGGUCAGA--AAUGUUAAACCAAUGAGCCUUUUUAUUAGCUUUAGAAUAAA-------- ...((.((-(((((((..--..))....))).)))))).......................-------- ( -7.30, z-score = 0.19, R) >droYak2.chr2R 266274 64 - 21139217 AUCCAGUC-GGGGUCACCAAAAUAUUGAGCUAAUAAGAAUUUGUAUUAGCCUUUUUGCAAAUGAA---- ((((....-.))))...(((((....(.(((((((........)))))))).)))))........---- ( -8.80, z-score = 0.46, R) >droAna3.scaffold_13417 6814960 62 - 6960332 AUCAUGUC-GGGGUCACC--AAUGUUUGGUCAUUGAGAUUUUUCAAUGACUUUUAUCCAAAAUAA---- ..(((...-.((....))--.)))...((((((((((....))))))))))..............---- ( -12.60, z-score = -0.69, R) >dp4.Unknown_group_284 5815 67 + 17202 AUCACGUC-GGGGUUACC-AAAUGUUAAGCUAUUAAGCGUUUUUAAUAAAUGUUGCAAUUUGAAGUUAA ...((((.-.((....))-..))))..(((((((.(((((((.....)))))))..)))....)))).. ( -8.30, z-score = 0.61, R) >droPer1.super_226 23616 67 - 38019 AUCACGUC-GGGGUCACC-AAAUGUUAAGCUAUUAAGCGUUUUUAAUAACUGUUGCAAUUUGAAGUUAA ...((((.-.((....))-..))))...(((....)))........(((((............))))). ( -8.50, z-score = 0.69, R) >droGri2.scaffold_14948 4364 59 - 44767 AUCACGUC-GGGGUCACCAGAGGUUUGCGCUAUCAG--CUUUUUGAUAGCUUUAAUUUCAAU------- ........-((.....)).((((((...((((((((--....))))))))...))))))...------- ( -15.40, z-score = -2.51, R) >droVir3.scaffold_12958 3369160 60 + 3547706 AUCACGUC-GGGGUCACC-AAAUGUUGCGCUAUUGAGCUUUUUCAAUAGCUUUCACAAAUAG------- ...((((.-.((....))-..))))...(((((((((....)))))))))............------- ( -14.40, z-score = -2.54, R) >droEre2.scaffold_4784 25734147 59 - 25762168 AUCACGUC-GGGGUCACC--AAUGUUACGAGGACUGACUGGUUUGACAGCCCUUUUCAAUAA------- ...((((.-((.....))--.))))...((((.(((.(......).))).))))........------- ( -10.10, z-score = 1.21, R) >consensus AUCACGUC_GGGGUCACC__AAUGUUAAGCUAUUGAGCUUUUUUAAUAGCUUUUGUAAAUAA_A_____ ...((((...((....))...))))...(((((((((....)))))))))................... ( -5.77 = -5.58 + -0.19)

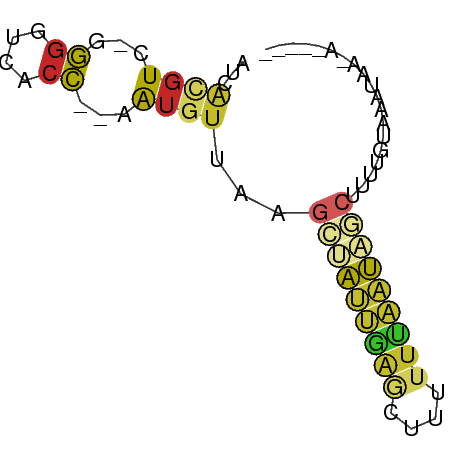
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:00:51 2011