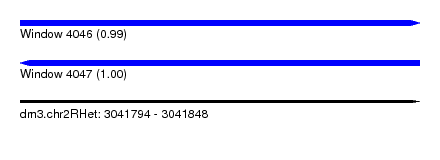
| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 3,041,794 – 3,041,848 |
| Length | 54 |
| Max. P | 0.999876 |

| Location | 3,041,794 – 3,041,848 |
|---|---|
| Length | 54 |
| Sequences | 4 |
| Columns | 55 |
| Reading direction | forward |
| Mean pairwise identity | 60.06 |
| Shannon entropy | 0.68267 |
| G+C content | 0.44572 |
| Mean single sequence MFE | -14.45 |
| Consensus MFE | -6.20 |
| Energy contribution | -5.45 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.986631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
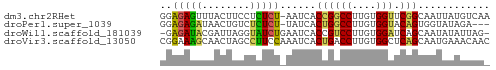
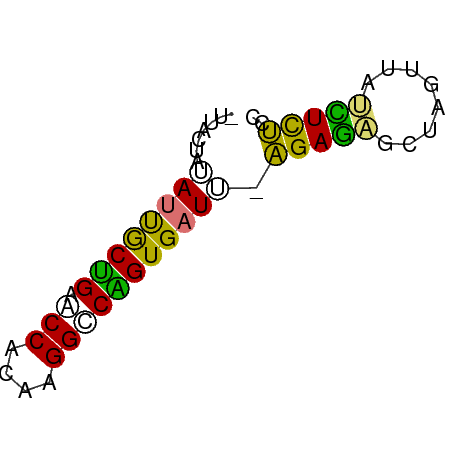
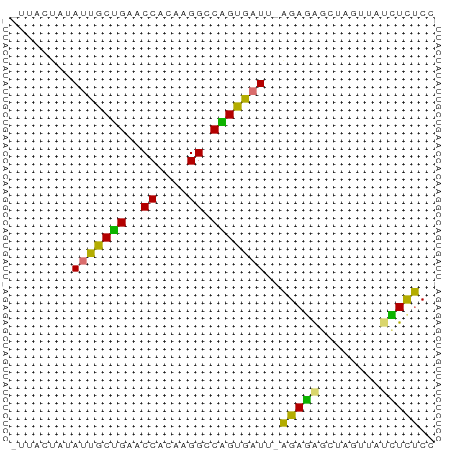
>dm3.chr2RHet 3041794 54 + 3288761 GGAGAGUUUACUUCCUCUCU-AAUCACCGGCCUUGUGGUUCGGCAAUUAUGUCAA ((((((........))))))-(((..((((((....)).))))..)))....... ( -13.30, z-score = -1.42, R) >droPer1.super_1039 3433 51 + 6997 GGAGAGAUAACUGUCUCUCU-UAUCACUGGCCUUGUGGUACAGUGGUAUAGA--- ((((((((....))))))))-(((((((((((....))).))))))))....--- ( -22.30, z-score = -3.88, R) >droWil1.scaffold_181039 452276 53 + 740211 -GAGAUACGAUUAGGUAUCUGAAUCACCGUCCUUGUGGAUCAGCAAUAUAUUAG- -.((((((......))))))........((((....))))..............- ( -9.30, z-score = -0.29, R) >droVir3.scaffold_13050 1180827 55 - 3426691 CGGAAAGCAACUAGCCUUCCAAAUCACUGACCUUGUGGCUCAGCAAUGAAACAAC .((((.((.....)).))))...(((((((((....)).))))...)))...... ( -12.90, z-score = -1.95, R) >consensus GGAGAGACAACUAGCUCUCU_AAUCACCGGCCUUGUGGUUCAGCAAUAAAGUAA_ ..(((((........)))))......((((((....))).)))............ ( -6.20 = -5.45 + -0.75)
| Location | 3,041,794 – 3,041,848 |
|---|---|
| Length | 54 |
| Sequences | 4 |
| Columns | 55 |
| Reading direction | reverse |
| Mean pairwise identity | 60.06 |
| Shannon entropy | 0.68267 |
| G+C content | 0.44572 |
| Mean single sequence MFE | -15.90 |
| Consensus MFE | -10.30 |
| Energy contribution | -8.93 |
| Covariance contribution | -1.38 |
| Combinations/Pair | 1.67 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.67 |
| SVM RNA-class probability | 0.999876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
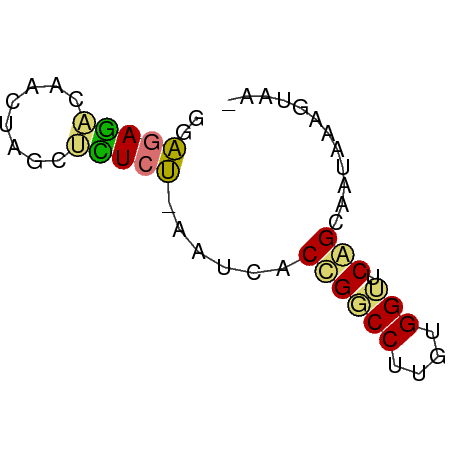
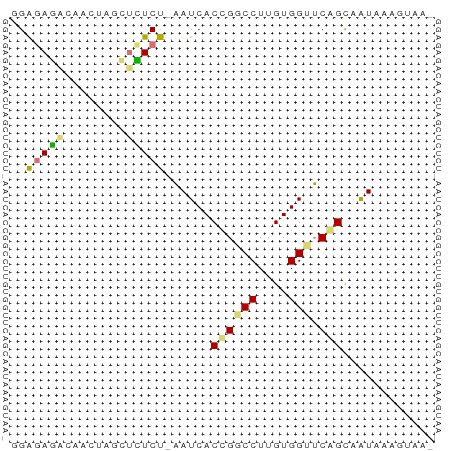
>dm3.chr2RHet 3041794 54 - 3288761 UUGACAUAAUUGCCGAACCACAAGGCCGGUGAUU-AGAGAGGAAGUAAACUCUCC ......((((..(((..((....)).)))..)))-)(((((........))))). ( -17.20, z-score = -3.65, R) >droPer1.super_1039 3433 51 - 6997 ---UCUAUACCACUGUACCACAAGGCCAGUGAUA-AGAGAGACAGUUAUCUCUCC ---.......(((((..((....)).)))))...-.((((((......)))))). ( -13.70, z-score = -2.42, R) >droWil1.scaffold_181039 452276 53 - 740211 -CUAAUAUAUUGCUGAUCCACAAGGACGGUGAUUCAGAUACCUAAUCGUAUCUC- -.......((..(((.(((....))))))..))..((((((......)))))).- ( -12.80, z-score = -2.20, R) >droVir3.scaffold_13050 1180827 55 + 3426691 GUUGUUUCAUUGCUGAGCCACAAGGUCAGUGAUUUGGAAGGCUAGUUGCUUUCCG ........((..((((.((....))))))..))..(((((((.....))))))). ( -19.90, z-score = -3.00, R) >consensus _UUACUAUAUUGCUGAACCACAAGGCCAGUGAUU_AGAGAGCUAGUUAUCUCUCC ........(((((((.(((....))))))))))..(((((........))))).. (-10.30 = -8.93 + -1.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:00:48 2011