
| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 2,874,810 – 2,874,880 |
| Length | 70 |
| Max. P | 0.536651 |

| Location | 2,874,810 – 2,874,880 |
|---|---|
| Length | 70 |
| Sequences | 5 |
| Columns | 74 |
| Reading direction | forward |
| Mean pairwise identity | 60.70 |
| Shannon entropy | 0.68601 |
| G+C content | 0.43545 |
| Mean single sequence MFE | -14.48 |
| Consensus MFE | -5.12 |
| Energy contribution | -5.80 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.536651 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
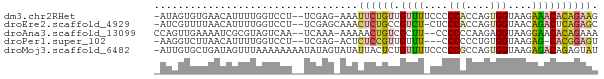
>dm3.chr2RHet 2874810 70 + 3288761 -AUAGUGUGAACAUUUUGGUCCU--UCGAG-AAAUUCUGUUGUUUUCCCCCACCAGUGGUAAGAAACACAGAAG -....((....))((((((....--)))))-)..((((((.(((((...(((....)))...))))))))))). ( -14.90, z-score = -1.06, R) >droEre2.scaffold_4929 4923718 70 - 26641161 -AUCGUUUUAACAUUUUGGUCCU--UCGAGCAAACUCUGCCGUCU-CUCCCACCAGUGGUAACAGACUCAGAGC -.............((((.((..--..)).))))(((((..((((-...(((....)))....)))).))))). ( -14.80, z-score = -1.05, R) >droAna3.scaffold_13099 1911989 69 - 3324263 CCAGUUGAAAAUCGCGUAGUCAA--UCAAA-AAAAACUGUCGCUU--CCCCCCAAGAGGUAAGGAACACAGAAA ...........(((((((((...--.....-....)))).)))((--((..((....))...))))....)).. ( -8.70, z-score = -0.02, R) >droPer1.super_102 69864 66 - 150666 -AAGGUCUUAACAUUUUGGUCCU--UCGAG-ACUCUCCGUUGUUU---CCCCCCUGUGGUAAGAG-CACGGAGU -..((((((..............--..)))-)))((((((.((((---...((....))...)))-))))))). ( -14.89, z-score = 0.20, R) >droMoj3.scaffold_6482 538196 73 + 2735782 -AUUGUGCUGAUAGUUUAAAAAAAAUAUAGUAUAUUACUCUGUUUUCCCCCGCCAGUGGUAAGAGACAGAGUAU -..(((((((...((((.....)))).))))))).(((((((((((...(((....)))...))))))))))). ( -19.10, z-score = -4.33, R) >consensus _AUGGUGUUAACAUUUUGGUCCU__UCGAG_AAACUCUGUUGUUU_CCCCCACCAGUGGUAAGAGACACAGAAU ..................................((((((.((((....(((....)))....)))))))))). ( -5.12 = -5.80 + 0.68)


| Location | 2,874,810 – 2,874,880 |
|---|---|
| Length | 70 |
| Sequences | 5 |
| Columns | 74 |
| Reading direction | reverse |
| Mean pairwise identity | 60.70 |
| Shannon entropy | 0.68601 |
| G+C content | 0.43545 |
| Mean single sequence MFE | -16.46 |
| Consensus MFE | -6.06 |
| Energy contribution | -7.18 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.535005 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2RHet 2874810 70 - 3288761 CUUCUGUGUUUCUUACCACUGGUGGGGGAAAACAACAGAAUUU-CUCGA--AGGACCAAAAUGUUCACACUAU- .((((((.((((((.(((....)))))))))...))))))...-.....--.((((......)))).......- ( -16.10, z-score = -0.80, R) >droEre2.scaffold_4929 4923718 70 + 26641161 GCUCUGAGUCUGUUACCACUGGUGGGAG-AGACGGCAGAGUUUGCUCGA--AGGACCAAAAUGUUAAAACGAU- ((((((.((((.((.(((....))))).-))))..))))))(((.((..--..)).)))..............- ( -21.10, z-score = -1.81, R) >droAna3.scaffold_13099 1911989 69 + 3324263 UUUCUGUGUUCCUUACCUCUUGGGGGG--AAGCGACAGUUUUU-UUUGA--UUGACUACGCGAUUUUCAACUGG ....(((.((((((.((....))))))--))))).(((((...-...((--(((......)))))...))))). ( -13.20, z-score = 0.04, R) >droPer1.super_102 69864 66 + 150666 ACUCCGUG-CUCUUACCACAGGGGGG---AAACAACGGAGAGU-CUCGA--AGGACCAAAAUGUUAAGACCUU- .((((((.-(((((.....)))))(.---...).)))))).((-((...--.)))).................- ( -16.20, z-score = 0.05, R) >droMoj3.scaffold_6482 538196 73 - 2735782 AUACUCUGUCUCUUACCACUGGCGGGGGAAAACAGAGUAAUAUACUAUAUUUUUUUUAAACUAUCAGCACAAU- .((((((((((((..((...))..)))))...)))))))..................................- ( -15.70, z-score = -3.11, R) >consensus AUUCUGUGUCUCUUACCACUGGGGGGGG_AAACAACAGAAUUU_CUCGA__AGGACCAAAAUGUUAACACCAU_ .((((((((((....(((....)))....))))).))))).................................. ( -6.06 = -7.18 + 1.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:00:45 2011