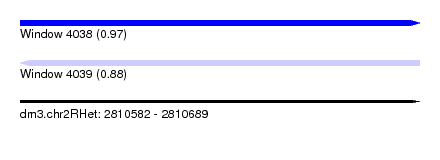
| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 2,810,582 – 2,810,689 |
| Length | 107 |
| Max. P | 0.965655 |

| Location | 2,810,582 – 2,810,689 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 52.64 |
| Shannon entropy | 0.78193 |
| G+C content | 0.52314 |
| Mean single sequence MFE | -31.62 |
| Consensus MFE | -10.25 |
| Energy contribution | -11.50 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.965655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2RHet 2810582 107 + 3288761 -------AUGCCGAAGAUCCCAUUGUACAUGAUUUUCCACAGGAUUGGUCAAAGUACCGAUCCUUGGAGAACACCUGUUGAAACUCGGUAUCUUUUUUGGCCACCUUCCGUAUC -------..(((((((((.((...(((((((.(((((((.(((((((((......))))))))))))))))))..)))....))..)).))))...)))))............. ( -30.20, z-score = -2.19, R) >dp4.Unknown_singleton_2768 1291 107 - 14987 -------AGUCGCAAUAUGCCGUCGUUCAUUGCGUUGCACAGUGUGGGGCCCAGCACCGACCCCUGCGGGACGCCUGCAAAUACCUCCUAUUGCUCCGUGCCAGCCUCGGUACC -------.(..(((((((((...(((.....)))..)))...((..((((((.(((........)))))).).))..)).........))))))..)(((((......))))). ( -31.30, z-score = 1.00, R) >droWil1.scaffold_181136 1555138 114 + 2313701 CAUUUCCAGUCGUAAGACAGUGUCAUACAUGACAUUCCUCAAUGCUGGCCCUAUCACCGUUCCUUGAGGUUCUCCGCCUGUGAUGGUGUGUUUCCGUGGUCCAGCUGCCGUAUC ........(((....)))(((((((....))))))).......((((((((...((((((..(..(.((....)).)..)..)))))).......).)).)))))......... ( -31.30, z-score = -1.33, R) >triCas2.chrUn_190 48519 104 - 65089 -------AAUCUAAGAACUCCAUCAUAAAGGAUGUUCCACAAGGUCGGCCCCAAUACCGAUCCUUGUGGAACUCCACUCGUCACCUC-CAUGACUUGUCCCUCGCCCAAGUC-- -------......................(((.(((((((((((((((........)))).))))))))))))))............-...((((((.........))))))-- ( -33.70, z-score = -5.17, R) >consensus _______AGUCGAAAGACACCAUCAUACAUGACGUUCCACAAGGUUGGCCCCAGCACCGAUCCUUGAGGAACUCCUCCUGAAACCUCGUAUUUCUUUGGGCCAGCCUCCGUAUC .....................(((......)))(((((((((((((((........))))).)))))))))).......................................... (-10.25 = -11.50 + 1.25)



| Location | 2,810,582 – 2,810,689 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 52.64 |
| Shannon entropy | 0.78193 |
| G+C content | 0.52314 |
| Mean single sequence MFE | -34.45 |
| Consensus MFE | -13.27 |
| Energy contribution | -15.03 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.879656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2RHet 2810582 107 - 3288761 GAUACGGAAGGUGGCCAAAAAAGAUACCGAGUUUCAACAGGUGUUCUCCAAGGAUCGGUACUUUGACCAAUCCUGUGGAAAAUCAUGUACAAUGGGAUCUUCGGCAU------- ....(.((((((..(((.....((((((..((....)).)))))).(((((((((.(((......))).))))).)))).............))).)))))).)...------- ( -27.80, z-score = -0.42, R) >dp4.Unknown_singleton_2768 1291 107 + 14987 GGUACCGAGGCUGGCACGGAGCAAUAGGAGGUAUUUGCAGGCGUCCCGCAGGGGUCGGUGCUGGGCCCCACACUGUGCAACGCAAUGAACGACGGCAUAUUGCGACU------- (((..(((.((((((((((.((....(((.((........)).))).)).((((((.......))))))...))))))..((.......)).))))...)))..)))------- ( -34.40, z-score = 1.41, R) >droWil1.scaffold_181136 1555138 114 - 2313701 GAUACGGCAGCUGGACCACGGAAACACACCAUCACAGGCGGAGAACCUCAAGGAACGGUGAUAGGGCCAGCAUUGAGGAAUGUCAUGUAUGACACUGUCUUACGACUGGAAAUG ....((((.(((((.((..(....).((((.(..(..(.((....)).)..)..).))))...)).)))))..(((((..(((((....)))))...))))).).)))...... ( -31.00, z-score = -0.96, R) >triCas2.chrUn_190 48519 104 + 65089 --GACUUGGGCGAGGGACAAGUCAUG-GAGGUGACGAGUGGAGUUCCACAAGGAUCGGUAUUGGGGCCGACCUUGUGGAACAUCCUUUAUGAUGGAGUUCUUAGAUU------- --..........((((((..((((((-(((....)....((((((((((((((.(((((......)))))))))))))))).)))))))))))...)))))).....------- ( -44.60, z-score = -4.61, R) >consensus GAUACGGAAGCUGGCCAAAAGAAAUACGAGGUAACAACAGGAGUUCCACAAGGAUCGGUACUGGGGCCAACACUGUGGAACAUCAUGUACGACGGAAUCUUACGACU_______ .......................................((((((((((((((...(((......)))..)))))))))))))).............................. (-13.27 = -15.03 + 1.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:00:41 2011