| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 2,649,483 – 2,649,548 |
| Length | 65 |
| Max. P | 0.957800 |

| Location | 2,649,483 – 2,649,548 |
|---|---|
| Length | 65 |
| Sequences | 4 |
| Columns | 66 |
| Reading direction | forward |
| Mean pairwise identity | 59.49 |
| Shannon entropy | 0.67380 |
| G+C content | 0.45268 |
| Mean single sequence MFE | -16.60 |
| Consensus MFE | -7.15 |
| Energy contribution | -7.28 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.957800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2RHet 2649483 65 + 3288761 CGGCUGCGGAUGUUUAUGUCUGUUGGAUUGGGUGCCUUCUUGAGUGGAUCACUUAUGGGUACUGA- ...(.((((((......)))))).).....(((((((...((((((...)))))).)))))))..- ( -19.60, z-score = -2.04, R) >droEre2.scaffold_4845 22460276 58 - 22589142 CGAUCGCGAAAGUUU--AUUAUUUCAGUUGGCUCGCCAAGUGGUCAAAU-GCAAAUGAUUA----- .((((((((((....--....))))..((((....))))))))))....-...........----- ( -13.80, z-score = -1.53, R) >droYak2.chr2L 1186444 64 - 22324452 CGUGGGCGGAAGUUUUUGUUAUUUCAGUUGGCUUGCCAAGUGAUAGAAU-GUUUACGU-ACGCAGA .....(((...((...((((...(((.((((....)))).)))...)))-)...))..-.)))... ( -16.30, z-score = -1.36, R) >droSec1.super_103 13908 64 - 108480 UGUGGGCGGAAGUUUAAGUUUUUUCAGUUGGCUCGCCAAGUGAUCGAAU-GUUUAGGGGACGUGG- ..(((((....))))).((((..(((.((((....)))).)))..))))-.....(....)....- ( -16.70, z-score = -1.50, R) >consensus CGUGGGCGGAAGUUUAUGUUAUUUCAGUUGGCUCGCCAAGUGAUCGAAU_GCUUAUGGGAAGUGG_ .....((....))....((((..(((.((((....)))).)))..))))................. ( -7.15 = -7.28 + 0.12)

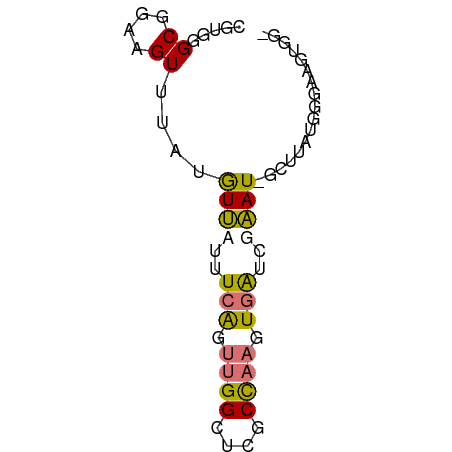
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:00:38 2011