| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 2,417,076 – 2,417,140 |
| Length | 64 |
| Max. P | 0.972078 |

| Location | 2,417,076 – 2,417,140 |
|---|---|
| Length | 64 |
| Sequences | 4 |
| Columns | 64 |
| Reading direction | forward |
| Mean pairwise identity | 57.52 |
| Shannon entropy | 0.68425 |
| G+C content | 0.43849 |
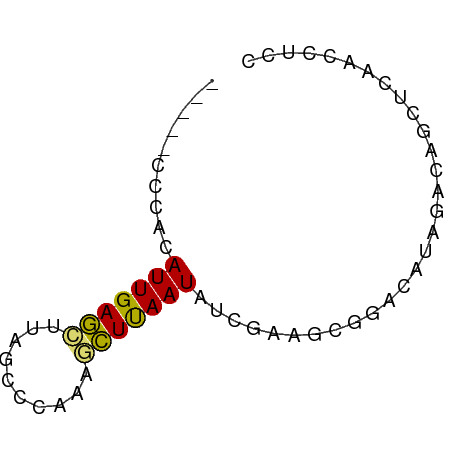
| Mean single sequence MFE | -11.53 |
| Consensus MFE | -4.91 |
| Energy contribution | -4.35 |
| Covariance contribution | -0.56 |
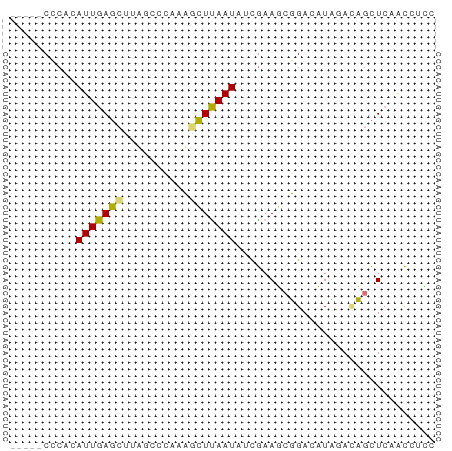
| Combinations/Pair | 1.43 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.972078 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
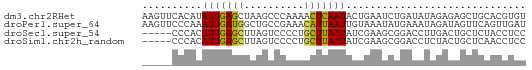
>dm3.chr2RHet 2417076 64 + 3288761 AAGUUCACAUAUUGAGCUAAGCCCAAAACUCAAUACUGAAUCUGAUAUAGAGAGCUGCACGUGU .(((((.(((((((((............))))))).))..((((...)))))))))........ ( -10.60, z-score = -0.38, R) >droPer1.super_64 209130 64 + 345335 AAGUUCCCAAAUUGAUGGCUGCCGAAACAUUAAUUGUAAAUAUGAAAUAGAUAGUUCAGUUGAU ..((..(((......)))..))......((((((((....(((.......)))...)))))))) ( -6.60, z-score = 0.34, R) >droSec1.super_54 91341 59 - 216317 -----CCCACAUUGAGCUUAGUCCCCUGCUUAAUAUCGAAGCGGACCUUGACUGCUCUACCUCC -----........((((..((((..((((((.......)))))).....))))))))....... ( -13.40, z-score = -2.71, R) >droSim1.chr2h_random 1328514 59 - 3178526 -----CCCACAUUGAGCUUAGUCCCCUGCUUAAUAUCGAAGCGGACCUCUACUGCUCAACCUCC -----......((((((.(((....((((((.......))))))....)))..))))))..... ( -15.50, z-score = -3.97, R) >consensus _____CCCACAUUGAGCUUAGCCCAAAGCUUAAUAUCGAAGCGGACAUAGACAGCUCAACCUCC ..........(((((((..........))))))).............................. ( -4.91 = -4.35 + -0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:00:33 2011