
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,119,000 – 2,119,109 |
| Length | 109 |
| Max. P | 0.644807 |

| Location | 2,119,000 – 2,119,109 |
|---|---|
| Length | 109 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 48.41 |
| Shannon entropy | 0.95569 |
| G+C content | 0.44163 |
| Mean single sequence MFE | -27.16 |
| Consensus MFE | -4.33 |
| Energy contribution | -5.01 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 2.21 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.16 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.644807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
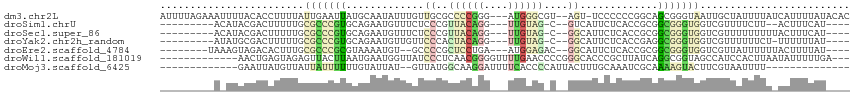
>dm3.chr2L 2119000 109 + 23011544 AUUUUAGAAAUUUUACACCUUUUAUUGAAUUAUGCAAUAUUUGUUGCGCCCCGGG---AUGGGCGU--AGU-UCCCCCCGGCAGCGGGUAAUUGCUAUUUUAUCAUUUUAUACAC ....(((.....((((.((..............(((((....)))))((.(((((---..(((...--...-.))))))))..))))))))...))).................. ( -23.90, z-score = -0.17, R) >droSim1.chrU 10533572 94 - 15797150 ---------ACAUACGACUUUUUGCGCCCGUGCAGAAUGUUUCUCCCGUUACAGG---UUGUAG-C--GUCAUUCUCACCGCGGCGGGUGGUCGUUUUCUU--ACUUUCAU---- ---------....((((((...(.((((((((.((((((.......((((((...---..))))-)--).)))))))).)).)))).).))))))......--........---- ( -28.70, z-score = -2.34, R) >droSec1.super_86 72737 96 + 128402 ---------ACAUACGACUUUUUGCGCCCGUGCAGAAUGUUUCUCCCGUUACAGG---UUGUAG-C--GGCAUUCUCACCGCGGCGGGUGGUCGUUUUUUUUUACUUUCAU---- ---------....((((((...(.((((((((.((((((......(((((((...---..))))-)--)))))))))).)).)))).).))))))................---- ( -32.20, z-score = -3.13, R) >droYak2.chr2h_random 1118208 95 + 3774259 ---------AUAUGCGACUUUUUGCGCCCGUGCAGAAUGUUGUUCCCACUACAGG---UUGUAG-C--GGCAUUCUCACCGAGGCGGGUGGUCGUUUUUUCU-UUUUUUAU---- ---------....((((((...(.((((((((.(((((((((((.(.((.....)---).).))-)--)))))))))).)).)))).).)))))).......-........---- ( -32.10, z-score = -3.03, R) >droEre2.scaffold_4784 1302520 96 - 25762168 --------UAAAGUAGACACUUUGCGCCCGCGUAAAAUGU--GCCCCGCUCCUGA---AUGGAGAC--GGCAUUCUCACCGCGGCGGGUGGUCGUUAUUUUUUACUUUUAU---- --------.((((((((((((.(.((((.(((......((--(((...((((...---..))))..--)))))......))))))).).))).)))......))))))...---- ( -31.20, z-score = -1.72, R) >droWil1.scaffold_181019 224223 99 - 324498 -------------AACUGAGUAGAGUUACUUAAUGAAUGGUUAUCCCUCAACGGGGUUUUGAACCCCGGGCACCCGCUUAUCAGGCGGUAGCCAUCCACUUAAUAUUUUUGA--- -------------...((((((....)))))).((.((((((((((((...((((((.....))))))(((....)))....))).))))))))).))..............--- ( -30.20, z-score = -1.79, R) >droMoj3.scaffold_6425 7454 86 - 194955 -------------GAAUUAUGUUAUUAUUUUUUGUAUUAU--GUUAUGGCAAGGAUUUUCACCCCAUUACUUUGCAAAUCGCAAAAGUACUUCGUAAUUUU-------------- -------------((((((((............(((((..--((.((((...((.......)))))).))(((((.....))))))))))..)))))))).-------------- ( -11.84, z-score = -0.38, R) >consensus _________A_AUAAGACAUUUUGCGCCCGUGCAGAAUGUUUGUCCCGCUACAGG___UUGUAG_C__GGCAUUCUCACCGCGGCGGGUGGUCGUUAUUUUU_ACUUUUAU____ ........................(((((((.............((..(((((......)))))....)).............)))))))......................... ( -4.33 = -5.01 + 0.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:10:13 2011