| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 2,342,193 – 2,342,249 |
| Length | 56 |
| Max. P | 0.997469 |

| Location | 2,342,193 – 2,342,249 |
|---|---|
| Length | 56 |
| Sequences | 5 |
| Columns | 59 |
| Reading direction | reverse |
| Mean pairwise identity | 58.32 |
| Shannon entropy | 0.73861 |
| G+C content | 0.42792 |
| Mean single sequence MFE | -13.33 |
| Consensus MFE | -7.70 |
| Energy contribution | -6.98 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.90 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.11 |
| SVM RNA-class probability | 0.997469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2RHet 2342193 56 - 3288761 UUCACAGUUUAAUGGCCUUAAGCUAUUAGAUGAAUCUCCAAAGCGCAAAAAAGCCG--- ......((((((((((.....))))))))))...........((........))..--- ( -10.00, z-score = -1.33, R) >dp4.Unknown_group_58 28533 56 + 69560 AGCUCAGUUUAGCGGUUUAAAACUGCUGAAUGAGUCUCCGAGGUGCAAAAAGGUCA--- .(((((.(((((((((.....)))))))))))))).....................--- ( -15.40, z-score = -1.43, R) >droPer1.super_79 151509 56 - 233179 GUCUCAAUUUGGCAGUCUGCAACUGCUGAAUGAGUCUCCGAGGCGUAAAAACGUCA--- (.((((.((..(((((.....)))))..)))))).).....(((((....))))).--- ( -20.00, z-score = -3.41, R) >droWil1.scaffold_181148 3150350 55 - 5435427 -UCUCAGUUUCGUAGUUUAACGCUACUGACUGAGUCUCGAAAGCGUAAGAAGGCAG--- -.(((((((..(((((.....))))).)))))))........((........))..--- ( -14.30, z-score = -0.89, R) >droGri2.scaffold_15252 722856 55 - 17193109 ----CAGUUUAAAAGUGUAAAACUCUCUAAAAACGCGCUUAAGCAGAAACGAUCAACUG ----..((((((..((((..............))))..))))))............... ( -6.94, z-score = -0.51, R) >consensus _UCUCAGUUUAGCAGUCUAAAACUACUGAAUGAGUCUCCAAAGCGCAAAAAAGCCA___ ......((((((((((.....))))))))))............................ ( -7.70 = -6.98 + -0.72)

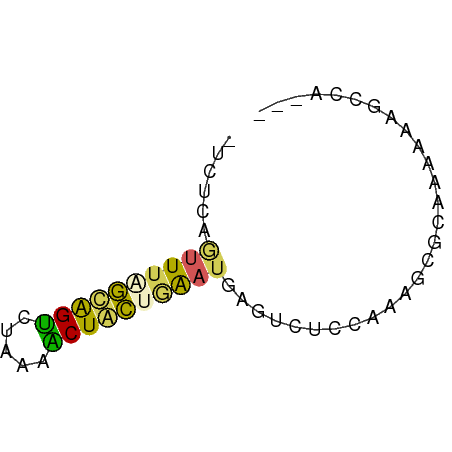
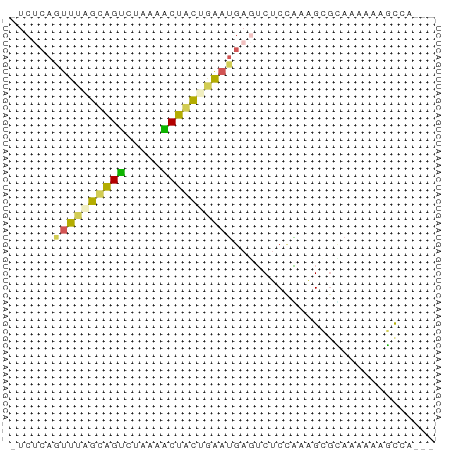
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:00:27 2011