| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 2,203,641 – 2,203,719 |
| Length | 78 |
| Max. P | 0.997932 |

| Location | 2,203,641 – 2,203,719 |
|---|---|
| Length | 78 |
| Sequences | 9 |
| Columns | 82 |
| Reading direction | reverse |
| Mean pairwise identity | 72.02 |
| Shannon entropy | 0.60349 |
| G+C content | 0.39229 |
| Mean single sequence MFE | -22.10 |
| Consensus MFE | -12.90 |
| Energy contribution | -11.91 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.81 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.997932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
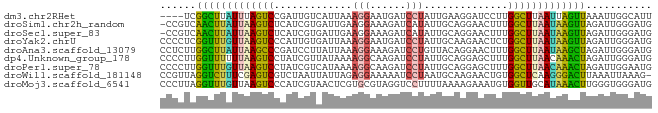
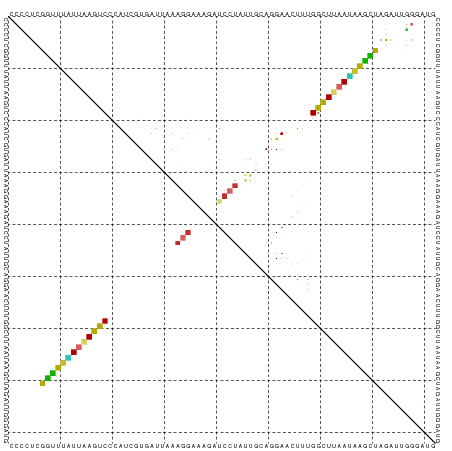
>dm3.chr2RHet 2203641 78 - 3288761 ----UCGGCUUAUUUAGUCCGAUUGUCAUUAAAGGAAUGAUCCUAUUGAAGGAUCCUUGGCUUAAUUAGUUAAAUUGGCAUU ----((((((.....)).)))).(((((..........((((((.....)))))).((((((.....))))))..))))).. ( -18.90, z-score = -1.60, R) >droSim1.chr2h_random 1802420 81 + 3178526 -CCGUCAACUUAUUAAGUCUCAUCGUGAUUGAAGGAAAGAUCAUAUUGCAGGAACUUUGGCUUAAUAAGUUAGAUUGGGAUG -((((((((((((((((((.((..((((((........))))))..))..........))))))))))))).)).))).... ( -21.00, z-score = -2.33, R) >droSec1.super_83 61957 81 + 109685 -CCGUCAACUUAUUAAGUCUCAUCGUGAUUGAAGGAAAGAUCAUAUUGCAGGAACUUUGGCUUAAUAAGUUAGAUUGGGAUG -((((((((((((((((((.((..((((((........))))))..))..........))))))))))))).)).))).... ( -21.00, z-score = -2.33, R) >droYak2.chrU 12959078 82 - 28119190 CCCCUCGGUUUGUUAAGUCCCAUUGUGAUUAAAGGAAUGAUCCUAUUGCAAGAACUCUGGCUUAAUAAGUUAGAUUGGGAUG ................(((((((((..((...((((....))))))..)))....(((((((.....))))))).)))))). ( -21.10, z-score = -1.73, R) >droAna3.scaffold_13079 412973 82 - 742402 CCUCUUGGCUUAUUAAGCCCGAUCCUUAUUAAAGGAAAGAUCCUGUUACAGGAACUUUGGCUUAAUAAGCUAGAUUGGGAUG (((.(((((((((((((((...(((((....)))))(((.(((((...))))).))).)))))))))))))))...)))... ( -33.20, z-score = -5.43, R) >dp4.Unknown_group_178 22519 82 + 23428 CCCCUUGGUUUUUUAAGUCCUAUCGUUAUAAAAGGCAAGAUCCUAUUGCAGGAGCUUUGGCUUAACAAACUAGAUUGGGAUG .((((((((((.(((((((.....(((......)))(((.((((.....)))).))).))))))).)))))))...)))... ( -20.10, z-score = -0.88, R) >droPer1.super_78 141533 82 - 255460 CCCCUUGGUUUGUUAAGUCCUAUCGUCAUAAAAGGCAAGAUCCUAUUGCAGGAGCUUUGGCUUAACAAACUAGAUUGGAAUG ((..(((((((((((((((.....(((......)))(((.((((.....)))).))).)))))))))))))))...)).... ( -25.20, z-score = -2.85, R) >droWil1.scaffold_181148 1701006 81 + 5435427 CCGUUAGGUCUUUCGAGUCGUCUAAUUAUUAGAGGAAAAAUCCUAAUGCAAGAACUGUGGCUCAAGGGACUUAAAUUAAAG- ...((((((((((.(((((.............((((....))))...(((.....)))))))).)))))))))).......- ( -20.70, z-score = -2.32, R) >droMoj3.scaffold_6541 2234887 82 + 2543558 CCCUUAGGUUUGUUAAGUCCCAUCGUAACUCGUGCGUAGGUCCUUUUAAAAGAAAUGUGGUUGCAUAAACUUGGGUGGGAUG (((((((((((...(((..((..((((.....))))..))..))).........((((....))))))))))))).)).... ( -17.70, z-score = -0.13, R) >consensus CCCCUCGGUUUAUUAAGUCCCAUCGUGAUUAAAGGAAAGAUCCUAUUGCAGGAACUUUGGCUUAAUAAGCUAGAUUGGGAUG ......(((((((((((((.............(((......)))..............)))))))))))))........... (-12.90 = -11.91 + -0.99)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:00:22 2011