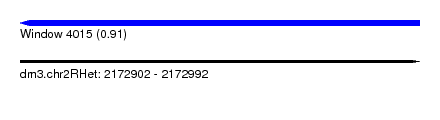
| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 2,172,902 – 2,172,992 |
| Length | 90 |
| Max. P | 0.906363 |

| Location | 2,172,902 – 2,172,992 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 53.54 |
| Shannon entropy | 0.63748 |
| G+C content | 0.53726 |
| Mean single sequence MFE | -33.10 |
| Consensus MFE | -10.80 |
| Energy contribution | -10.60 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.906363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2RHet 2172902 90 - 3288761 ---UGAGGAAGAAG--CCGGCG--GAUGGUGAUCUGGGCAUCUCCAGAUCAGUAACGC-----CGAUGAUGGAGCCGAUCAUUUAAUGGUCGGGAGUACAGG ---...........--.(((((--..((.(((((((((....))))))))).)).)))-----)).........((((((((...))))))))......... ( -33.60, z-score = -2.59, R) >droYak2.chr3R 144189 94 - 28832112 UGGUGAAGACGGAGACCUAACGUUGACAGUCAACCCGUCAAAUGGAGAUGAGCCAUGC-----CACUGAGGGUGCUG-CAGCUCAAUCUUCAUAAAAGCG-- ..((...(((((.(......)((((.....)))))))))..((((((((((((..(((-----(((.....)))..)-)))))).))))))))....)).-- ( -27.70, z-score = -1.23, R) >dp4.Unknown_group_70 2352 97 - 32930 -AAGGAAGACGGUGACCUGACGCUGGUAGUCAACCCGUCAGGUGCAAGUGAGCCAGCCACCCAUGCUGAGUGUGCUG-CAGCUCAACUUGCAUAAAAGC--- -.............((((((((..(((.....)))))))))))(((((((((((((((((.(.....).))).))))-..)))).))))))........--- ( -38.00, z-score = -2.49, R) >consensus ___UGAAGACGGAGACCUGACG_UGAUAGUCAACCCGUCAAAUGCAGAUGAGCCACGC_____CGCUGAGGGUGCUG_CAGCUCAAUCUUCAUAAAAGC___ .......(((((....(((.......))).....)))))..((((((((((((...........................)))).))))))))......... (-10.80 = -10.60 + -0.21)

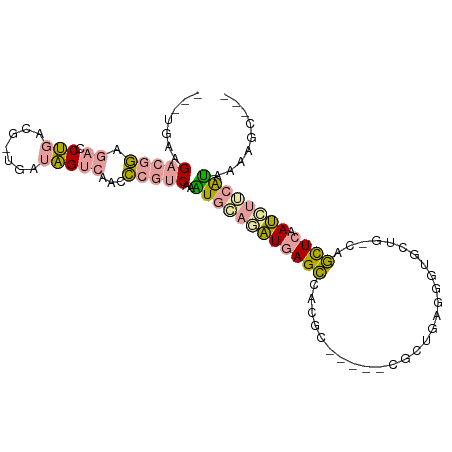
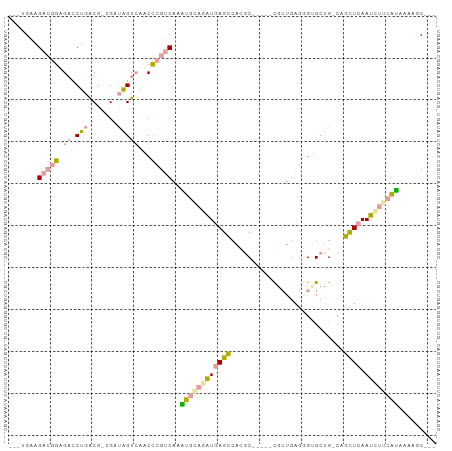
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:00:22 2011