
| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 2,169,205 – 2,169,309 |
| Length | 104 |
| Max. P | 0.527941 |

| Location | 2,169,205 – 2,169,309 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 63.16 |
| Shannon entropy | 0.75255 |
| G+C content | 0.49064 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -11.68 |
| Energy contribution | -11.77 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.42 |
| Mean z-score | -0.50 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.527941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
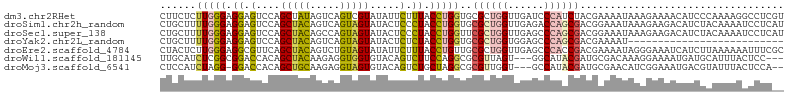
>dm3.chr2RHet 2169205 104 + 3288761 CUUCUCUUGGGAGGAGUCCAGCUAUAGUCAGUCGUAUAUUCUUUACCUGGUGCGCUGGUUGAUCCCAUCUACGAAAAUAAAGAAAACAUCCCAAAAGGCCUCGU ..(((.(((((((((..(((((....(((((..(((.......))))))))..)))))....)))..(((..........))).....)))))).)))...... ( -22.60, z-score = 0.07, R) >droSim1.chr2h_random 1209282 104 + 3178526 CUGCUUUUGGGAGGAGUCCAGCUACAGUCAGUAGUAUACUCCCUACCUGGUGCGCUGGUUGAGACCAGCGACGGAAAUAAAGAAGACAUCUACAAAAUCCUCAU ..((..(..((.(((((...(((((.....)))))..)))))...))..).))((((((....))))))((.(((.....(((.....)))......))))).. ( -30.60, z-score = -1.39, R) >droSec1.super_138 34490 104 + 55495 CUGCUUUUGGGAGGAGUCCAGCUACAGCCAGUAGUAUACUCCCUACCUGGUUCGCUGGUUGAGCCCAGCGACGGAAAUAAAGAAGACAUCUACAAAAUCCUCAU ..........(((((.(((.......((((((((........))).)))))(((((((......))))))).))).....(((.....)))......))))).. ( -29.80, z-score = -1.17, R) >droYak2.chr2L_random 2306275 78 - 4064425 CUGCUUUUGGGAGGAGUCCAGCUACAGUCAGUAGUAUACUCUCUACCUGGUGCGCUGGUGGAGCCCAGCGACGAAAAU-------------------------- ..((..(..((((((((...(((((.....)))))..)))).)).))..).))(((((......))))).........-------------------------- ( -25.60, z-score = -0.67, R) >droEre2.scaffold_4784 23384631 104 + 25762168 CUACUCUUGGGAGGCGUUCAGCUACAGUCUGUAGUAUAUUCUUUACCUGUUGCGCUGGUUGAGCCCACCGACGAAAAUAGGGAAAUCAUCUUAAAAAAUUUCGC ......((((..(((..(((((.((((...((((........))))))))...)))))....)))..))))(((((.(((((......))))).....))))). ( -22.80, z-score = 0.17, R) >droWil1.scaffold_181145 1389721 98 - 2682060 UUGCAUCUCGGCGGACCACAGCUACAAGAGGUGGUGUACAGUCUUCCAGGCGCGUUAGU---GGCAUACGAUGCGACAAAGGAAAAUGAUGCAUUUACUCC--- .((((((..((((.(((((..(.....)..))))).)...)))((((..(((((((.((---(...))))))))).)...))))...))))))........--- ( -26.70, z-score = -0.16, R) >droMoj3.scaffold_6541 592603 98 - 2543558 CUCCAUCUAGG-GGACCACAGCUGCAAGAGGUAGUGUACAGUCUGCUAGGCGCGUUGGU---GCCAUACGAUGCGAACAUCGGAAAUGACGUAUUUACUCCA-- .(((.(((((.-((((....(((((.....))))).....)))).)))))(((((((..---......)))))))......)))..................-- ( -29.30, z-score = -0.36, R) >consensus CUGCUUUUGGGAGGAGUCCAGCUACAGUCAGUAGUAUACUCUCUACCUGGUGCGCUGGUUGAGCCCAGCGACGAAAAUAAAGAAAACAUCUAAAAAACCCUC__ ......(((((.((.(....(((((.....))))).....).)).)))))..((((((......)))))).................................. (-11.68 = -11.77 + 0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:00:21 2011