| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 2,167,200 – 2,167,314 |
| Length | 114 |
| Max. P | 0.731409 |

| Location | 2,167,200 – 2,167,314 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 57.60 |
| Shannon entropy | 0.57728 |
| G+C content | 0.44460 |
| Mean single sequence MFE | -28.77 |
| Consensus MFE | -11.04 |
| Energy contribution | -12.60 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.731409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
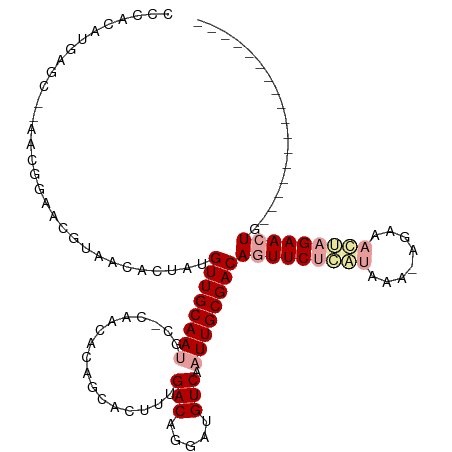
>dm3.chr2RHet 2167200 114 + 3288761 CCCA-GCGAUCUCAAGUGAUCACUACACUUUGUUGCAAUGC-CAACUCAUCA-UUGGACAGGAUGUCAAUUGCGACAGUUCGGAUGAA---CAACUGGAACUGCAAGAAUGCUUAUGUUG .(((-(.((((......)))).........(((((((((..-.....((...-.))(((.....))).)))))))))((((....)))---)..))))(((.(((....)))....))). ( -26.60, z-score = 0.15, R) >droEre2.scaffold_4845 1029633 102 + 22589142 CGCACAUAAGCCGAACGGAACGUACCAUUCUGUUGCAAUGCAUUCUGUUGCACUUUGACAGGAUGUCGAUUGCGACAUUUCUCGCCGA-AGAAACUAGAACUG----------------- .((......)).(((.((......)).)))(((((((((((((((((((.......))))))))))..)))))))))(((((......-))))).........----------------- ( -26.60, z-score = -1.36, R) >droYak2.chr2h_random 3013004 101 + 3774259 CUCACUUGAGC--AAUGGAAUGUAUUACAAUGUUGCAAGUGCCGACACAGCACUUUGACAGGAUGUCCAUUGCGACAGUUCUUAUAAACAGAAAUGAGAACUG----------------- .........((--((((((...((((....(((((.((((((.......))))))))))).))))))))))))..((((((((((........))))))))))----------------- ( -33.10, z-score = -4.00, R) >consensus CCCACAUGAGC__AACGGAACGUAACACUAUGUUGCAAUGC_CAACACAGCACUUUGACAGGAUGUCAAUUGCGACAGUUCUCAUAAA_AGAAACUAGAACUG_________________ ...............................((((((((.................(((.....))).))))))))(((((((((........))))))))).................. (-11.04 = -12.60 + 1.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:00:20 2011