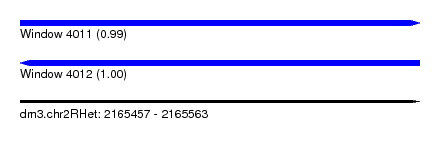
| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 2,165,457 – 2,165,563 |
| Length | 106 |
| Max. P | 0.997131 |

| Location | 2,165,457 – 2,165,563 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 59.75 |
| Shannon entropy | 0.56619 |
| G+C content | 0.57547 |
| Mean single sequence MFE | -44.60 |
| Consensus MFE | -19.00 |
| Energy contribution | -18.97 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.994984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
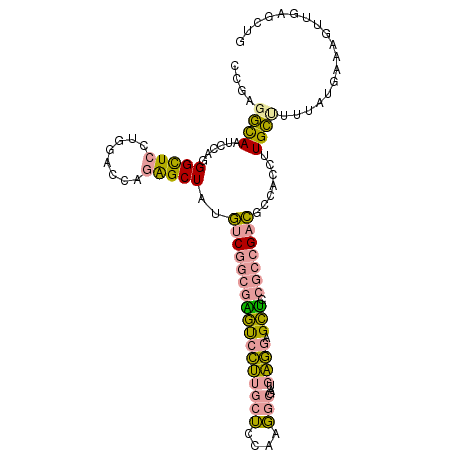
>dm3.chr2RHet 2165457 106 + 3288761 CCUGGUUAUACCAGGGUUCCUGUACCAGGGCUAGCCCAAGAUGUUGCUUGGUGAACAUCCUUGCCAGCACGGCCGAGGCCGCCUUGGCGCGAUGGAUGUUCACCUG (((((.....)))))............(((....)))............(((((((((((.((((((..((((....))))..))))))....))))))))))).. ( -49.20, z-score = -3.46, R) >droYak2.chrU 16962128 106 - 28119190 CCGAGGCAAUCCAUGGCUCUUGGACCAAAGCUAUGUCGGCGGACCCUUGCUCCAGGGCGAUGAGGAGUUCCGCCGACGCGAUCUUGCUUUUAUAAAGAUUGAGCUG (((((((........)).))))).....((((..(((((((((((((((((....)))...)))).).))))))))).(((((((.........))))))))))). ( -43.50, z-score = -3.08, R) >dp4.Unknown_group_70 2267 106 + 32930 CAGUCGCAAUCCAGGGCUCCUGGACUAGAGCUAUGUCGGCGAGUCCUUGCUCCAUGGCGAUGAGGAGCUCCGCCGACGCUACCUUGCUUUUAUGCAAGUUGAGCUG .(((.....((((((...)))))).....)))..(((((((((((((((((....)))...))))).)).)))))))(((..(((((......)))))...))).. ( -41.10, z-score = -1.61, R) >consensus CCGAGGCAAUCCAGGGCUCCUGGACCAGAGCUAUGUCGGCGAGUCCUUGCUCCAAGGCGAUGAGGAGCUCCGCCGACGCCACCUUGCUUUUAUGAAAGUUGAGCUG ....((((......(((((........)))))..(((((((((((((((((....)))...)))).))).))))))).......)))).................. (-19.00 = -18.97 + -0.04)

| Location | 2,165,457 – 2,165,563 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 59.75 |
| Shannon entropy | 0.56619 |
| G+C content | 0.57547 |
| Mean single sequence MFE | -42.70 |
| Consensus MFE | -21.68 |
| Energy contribution | -18.82 |
| Covariance contribution | -2.86 |
| Combinations/Pair | 1.55 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.997131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
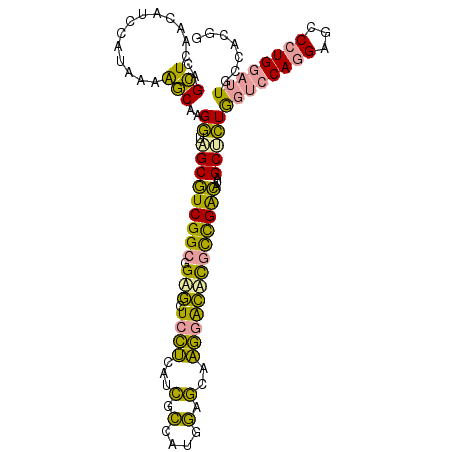
>dm3.chr2RHet 2165457 106 - 3288761 CAGGUGAACAUCCAUCGCGCCAAGGCGGCCUCGGCCGUGCUGGCAAGGAUGUUCACCAAGCAACAUCUUGGGCUAGCCCUGGUACAGGAACCCUGGUAUAACCAGG ..(((((((((((.....((((..(((((....)))))..))))..)))))))))))........(((((..(((....)))..)))))..(((((.....))))) ( -51.70, z-score = -4.36, R) >droYak2.chrU 16962128 106 + 28119190 CAGCUCAAUCUUUAUAAAAGCAAGAUCGCGUCGGCGGAACUCCUCAUCGCCCUGGAGCAAGGGUCCGCCGACAUAGCUUUGGUCCAAGAGCCAUGGAUUGCCUCGG .....((((((........((......))(((((((((.(.(((..((......))...)))))))))))))...(((((......)))))...))))))...... ( -32.80, z-score = -0.42, R) >dp4.Unknown_group_70 2267 106 - 32930 CAGCUCAACUUGCAUAAAAGCAAGGUAGCGUCGGCGGAGCUCCUCAUCGCCAUGGAGCAAGGACUCGCCGACAUAGCUCUAGUCCAGGAGCCCUGGAUUGCGACUG ..(((...(((((......)))))..)))(((((((.((.((((..((......))...))))))))))))).(((.(((((((((((...))))))))).))))) ( -43.60, z-score = -2.78, R) >consensus CAGCUCAACAUCCAUAAAAGCAAGGUAGCGUCGGCGGAGCUCCUCAUCGCCAUGGAGCAAGGACACGCCGACAUAGCUCUGGUCCAGGAGCCCUGGAUUGCCACGG ..(((.............)))..((.(((((((((.(((.((((...(.(....).)..)))))))))))))...)))))((((((((...))))))))....... (-21.68 = -18.82 + -2.86)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:00:19 2011