
| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 1,964,584 – 1,964,683 |
| Length | 99 |
| Max. P | 0.776000 |

| Location | 1,964,584 – 1,964,683 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 50.66 |
| Shannon entropy | 0.88232 |
| G+C content | 0.45177 |
| Mean single sequence MFE | -24.39 |
| Consensus MFE | -6.27 |
| Energy contribution | -8.11 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.70 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.776000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
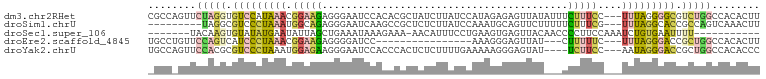
>dm3.chr2RHet 1964584 99 - 3288761 CGCCAGUUCUAGGUGUCCAUAAACGGAAGAGGGAAUCCACACGCUAUCUUAUCCAUAGAGAGUUAUAUUUCUUUCC---UUUAGGGGCGUCUGGCCACACUU .(((((.....((..(((.....(....)..)))..))..(((((..((((.....((((((......))))))..---..))))))))))))))....... ( -24.10, z-score = -0.31, R) >droSim1.chrU 4317248 90 - 15797150 ---------UAGGCGUCCCUAAAUGGCAGAGGGAAUCAAGCCGCUCUCUUAUCCAAAUGCAGUUCUUUUUCUUUCG---UUUAGGCACCGCCAGUCAAACUU ---------..((((..(((((((((.(((((((....(((.((..............)).))).))))))).)))---))))))...)))).......... ( -23.74, z-score = -1.81, R) >droSec1.super_106 27657 83 + 90575 -------UACAAGUGUAUAUGAAUAUUAGCUGAAAUAAAGAAA-AACAUUUCCUGAAGUGAGUUACAACCCCUUCCAAAUCUGUGAAUUUU----------- -------((((..(............(((((.(......((((-....))))......).))))).............)..))))......----------- ( -4.61, z-score = 1.46, R) >droEre2.scaffold_4845 21324862 80 + 22589142 UGCCUGUUCCAGUCAUCCCUAAACGGAAGAGGGGAUCC----------------AAAGGGAGUUAU---CUUUUUC---UUUAGGGACCGCUGGCCACACUU ....(((.(((((..((((((((.((((((((...(((----------------....)))....)---)))))))---))))))))..)))))..)))... ( -30.20, z-score = -2.34, R) >droYak2.chrU 22898855 95 - 28119190 UGCCAGUUCCACGCGUCCCUAAAUGGAGAAGGGAAUCCACCCACUCUCUUUUGAAAAAGGGAGUAU----UCUUCC---AAUAGGGACCGCUGGCCACACCC .(((((.....((.(((((((..((((((((((......)))(((((((((....))))))))).)----)).)))---).))))))))))))))....... ( -39.30, z-score = -4.50, R) >consensus _GCC_GUUCCAGGCGUCCCUAAAUGGAAGAGGGAAUCCACACACUAUCUUAUCCAAAGGGAGUUAU___CCUUUCC___UUUAGGGACCGCUGGCCACACUU ........(((((.(((((((((.....((((((...................................))))))....))))))))).)))))........ ( -6.27 = -8.11 + 1.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:00:13 2011