| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 1,940,365 – 1,940,419 |
| Length | 54 |
| Max. P | 0.943837 |

| Location | 1,940,365 – 1,940,419 |
|---|---|
| Length | 54 |
| Sequences | 4 |
| Columns | 57 |
| Reading direction | forward |
| Mean pairwise identity | 68.17 |
| Shannon entropy | 0.51819 |
| G+C content | 0.36606 |
| Mean single sequence MFE | -5.25 |
| Consensus MFE | -4.47 |
| Energy contribution | -4.72 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.79 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.904042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
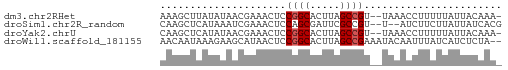
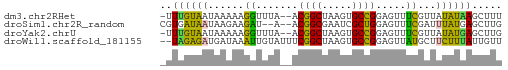
>dm3.chr2RHet 1940365 54 + 3288761 AAAGCUUAUAUAACGAAACUCCGGCACUUAGCCGU--UAAACCUUUUUAUUACAAA- .....................((((.....)))).--...................- ( -5.90, z-score = -0.80, R) >droSim1.chr2R_random 2685591 53 - 2996586 CAAGCUCAUAAAUCGAAACUCCAGCGAUUCGCCGU--U--AUCUUCUUAUUAUCACG .(((...((((..((((.(......).))))...)--)--))...)))......... ( -2.80, z-score = 0.52, R) >droYak2.chrU 1939929 54 + 28119190 CAAGCUCAUAUAACGAAACUCCGGCACUUAGCCGU--UAAACCUUUUUAUUACAAA- .....................((((.....)))).--...................- ( -5.90, z-score = -1.11, R) >droWil1.scaffold_181155 227040 55 - 3442787 AACAAUAAAGAAGCAUAACUCCGGCACUUAGCCGAAAUACAAUUUAUCAUCUCUA-- .....................((((.....)))).....................-- ( -6.40, z-score = -1.76, R) >consensus AAAGCUCAUAAAACGAAACUCCGGCACUUAGCCGU__UAAACCUUUUUAUUACAAA_ .....................((((.....))))....................... ( -4.47 = -4.72 + 0.25)
| Location | 1,940,365 – 1,940,419 |
|---|---|
| Length | 54 |
| Sequences | 4 |
| Columns | 57 |
| Reading direction | reverse |
| Mean pairwise identity | 68.17 |
| Shannon entropy | 0.51819 |
| G+C content | 0.36606 |
| Mean single sequence MFE | -11.23 |
| Consensus MFE | -8.07 |
| Energy contribution | -7.70 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.943837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
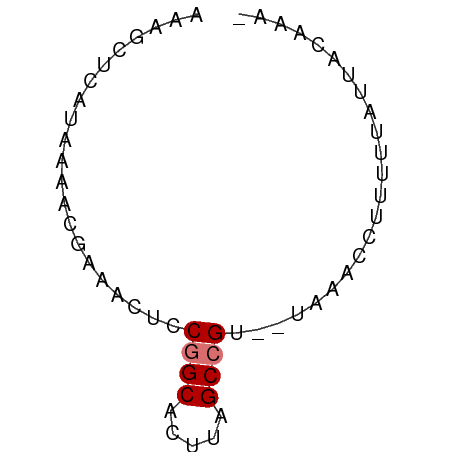
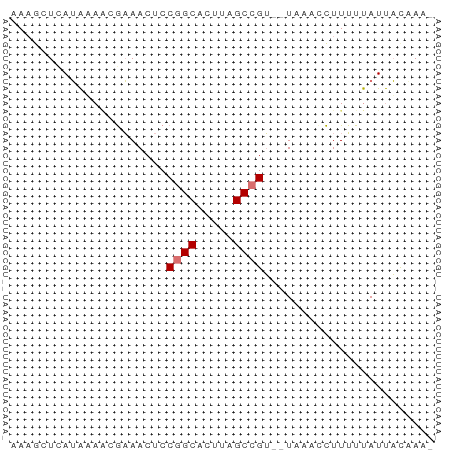
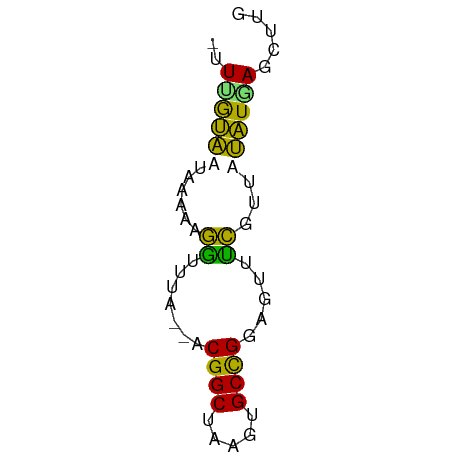
>dm3.chr2RHet 1940365 54 - 3288761 -UUUGUAAUAAAAAGGUUUA--ACGGCUAAGUGCCGGAGUUUCGUUAUAUAAGCUUU -...........((((((((--.((((.....))))(.....)......)))))))) ( -10.20, z-score = -0.84, R) >droSim1.chr2R_random 2685591 53 + 2996586 CGUGAUAAUAAGAAGAU--A--ACGGCGAAUCGCUGGAGUUUCGAUUUAUGAGCUUG .((.((((...((((..--.--.(((((...)))))...))))...))))..))... ( -10.40, z-score = -0.75, R) >droYak2.chrU 1939929 54 - 28119190 -UUUGUAAUAAAAAGGUUUA--ACGGCUAAGUGCCGGAGUUUCGUUAUAUGAGCUUG -...................--.((((.....))))((((((........)))))). ( -10.10, z-score = -0.69, R) >droWil1.scaffold_181155 227040 55 + 3442787 --UAGAGAUGAUAAAUUGUAUUUCGGCUAAGUGCCGGAGUUAUGCUUCUUUAUUGUU --....((..(((((..((((((((((.....))))))...))))...)))))..)) ( -14.20, z-score = -2.54, R) >consensus _UUUGUAAUAAAAAGGUUUA__ACGGCUAAGUGCCGGAGUUUCGUUAUAUGAGCUUG .......................((((.....))))((((((........)))))). ( -8.07 = -7.70 + -0.37)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:00:12 2011