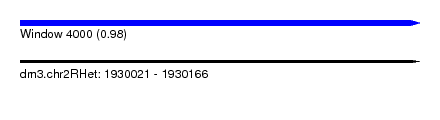
| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 1,930,021 – 1,930,166 |
| Length | 145 |
| Max. P | 0.984849 |

| Location | 1,930,021 – 1,930,166 |
|---|---|
| Length | 145 |
| Sequences | 6 |
| Columns | 177 |
| Reading direction | forward |
| Mean pairwise identity | 62.01 |
| Shannon entropy | 0.64085 |
| G+C content | 0.52156 |
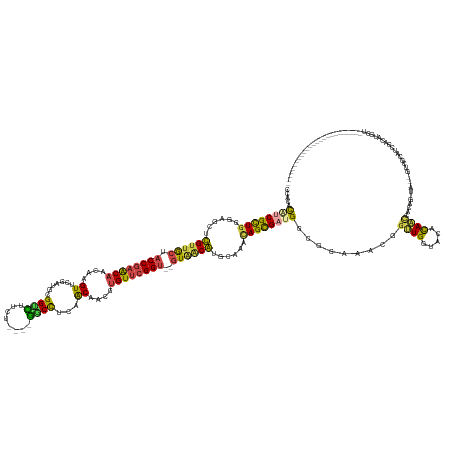
| Mean single sequence MFE | -50.58 |
| Consensus MFE | -25.76 |
| Energy contribution | -24.47 |
| Covariance contribution | -1.30 |
| Combinations/Pair | 1.61 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.984849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
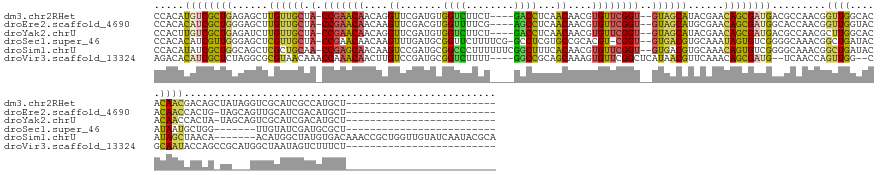
>dm3.chr2RHet 1930021 145 + 3288761 CCACAUGUCGCUGAGAGCUUGUUGCUA-CCGAACAACAGGUUCGAUGUGGUCUUCU----GACCUCAACAACGUGUUCGGU--GUAGCAUACGAACAGCGAUGACGCCAACGGUUGGCACACAACGACAGCUAUAGGUCGCAUCGCCAUGCU------------------------- ...((((.(((((...(..((((((.(-(((((((....(((.(.((.((((....----)))).)).)))).))))))))--))))))..)...))))((((..((((.....))))......((((........))))))))).))))..------------------------- ( -51.20, z-score = -1.75, R) >droEre2.scaffold_4690 760076 144 - 18748788 CCACACAUCGCUGGGAGCUUGUUGCUA-CCGAACAACAAGUUCGACGUGGUUUUCG----AGCCUCAACAACGUGUUCGGU--GUAGCAUGCGAACAGCGAUGGCACCAACGGUUGGUACACAACCACUG-UAGCAGUUGCAUCGACAUGCU------------------------- ..(((((((((((...((.((((((.(-(((((((....(((.(..(.((((....----)))).)..)))).))))))))--)))))).))...))))))))........(((((.....)))))..))-)((((((((...)))).))))------------------------- ( -55.40, z-score = -2.75, R) >droYak2.chrU 26149422 144 - 28119190 CCACUUGUCGCUGAGAUCUUGUUGCUA-CCGAACAACAGGUUCGAUGUGGUCUUCU----GACCUCAACAACGUGUUCGGU--GUAGCAUACGAACAGCGAUGACGCCAACGCUUGGCACACAACCACUA-UAGCAGUCGCAUCGACAUGCU------------------------- ....(..((((((......((((((.(-(((((((....(((.(.((.((((....----)))).)).)))).))))))))--))))))......))))))..).((((.....))))............-.((((((((...)))).))))------------------------- ( -51.30, z-score = -2.99, R) >droSec1.super_46 172757 140 + 322281 CCACACAUCGUUGGGAGCUCGUUGCUA-CCGAACAACAAGUUUGAUGCGGUUCUUUUCG-GCCUCGUGCCGCACGU-CGGU--GUGACGUGCAAAUAGUGUCGGGGCAAACGGCUGAUACAUAAUGCUGG-------UUGUAUCGAUGCGCU------------------------- ...(.(((((((.(((((.....))).-)).))).........(((((((.........-(((((((((.((((((-(...--..))))))).....))).))))))...((((...........)))).-------))))))))))).)..------------------------- ( -46.10, z-score = 0.17, R) >droSim1.chrU 13678307 167 - 15797150 CCACAUAUCGCUGGCAGCUCGCUGCAA-CCGAGCAACAAGUCCGAUGCGGCCCUUUUUUCGGCUUUCACAACGUGUUCGGU--GUGACGUGCAAACAGUGUCGGGGCAAACGGCUGAUACAUAGCUAACA-------ACAUGGCUAUGUGACAAACCGCUGGUUGUAUCAAUACGCA ...((..((((..((((....)))).(-(((((((....((..((...((((........)))).))...)).))))))))--))))..))......((((.(..((((.((((((.((((((((((...-------...)))))))))).))....)))).))))..).))))... ( -53.10, z-score = -0.12, R) >droVir3.scaffold_13324 464977 144 + 2960039 AGACACAUCGCUCUAGGCGCGUAACAAACCAAACAACUUGUCCGAUGCGGUCUUUU----GGCCGCAGCAAAGUGUUCGGCUCAUAACGUUCAAACAGCGAUG--UCAACCAGUUGG--CGCAAUACCAGCCGCAUGGCUAAUAGUCUUUCU------------------------- ((((((((((((....(.((((......((.((((..((((....(((((((....----)))))))))))..)))).))......)))).)....)))))))--)...(((..(((--(.........))))..)))......))))....------------------------- ( -46.40, z-score = -2.49, R) >consensus CCACACAUCGCUGGGAGCUCGUUGCUA_CCGAACAACAAGUUCGAUGCGGUCUUCU____GGCCUCAACAACGUGUUCGGU__GUAACAUGCAAACAGCGAUGGCGCAAACGGUUGGUACACAACCACAG_UA___GUCGCAUCGACAUGCU_________________________ .....((((((((......((((((...(((((((....((.......((((........))))...))....)))))))...))))))......)))))))).........((((.....)))).................................................... (-25.76 = -24.47 + -1.30)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:00:10 2011