| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 1,766,988 – 1,767,133 |
| Length | 145 |
| Max. P | 0.699128 |

| Location | 1,766,988 – 1,767,133 |
|---|---|
| Length | 145 |
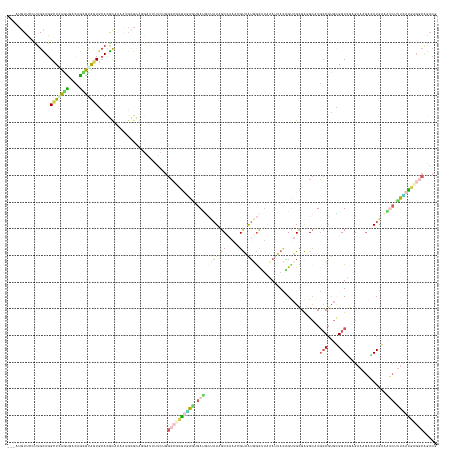
| Sequences | 5 |
| Columns | 161 |
| Reading direction | forward |
| Mean pairwise identity | 56.53 |
| Shannon entropy | 0.79244 |
| G+C content | 0.48162 |
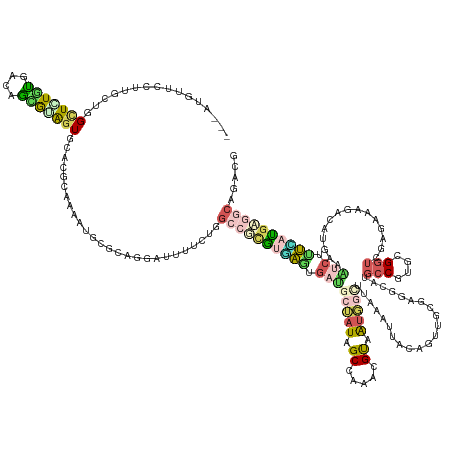
| Mean single sequence MFE | -51.86 |
| Consensus MFE | -12.96 |
| Energy contribution | -14.40 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.83 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.699128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2RHet 1766988 145 + 3288761 ---AAAUUACUUGUUGGCUAUGUGAUAGAAUAGUGCAUACCA-------------UUAUGGCCGUACAACUGAUGACCUAGCAAAGGGUAAAAAUCUAAAUUACUGUUGUGAUGCUUGCCUUGUGGUUGCGAACGAGAUGAAAUCGUUUAUGCGCCAAACU ---..........(((((...(..((...((((.(((...((-------------(((..((.(((.....(((.((((......))))....))).....))).))..)))))..))).)))).))..)((((((.......))))))....)))))... ( -37.00, z-score = -1.69, R) >dp4.Unknown_group_407 737 158 + 16403 ---AUGUCCCCUGCUGGCUCUGCGACAGCGUAGUGAACGCAAAAUGCGCUGGAUUUUCUGGCCUCGUGAGUGAUGCUAUAGCCAAACGUAAUGGCUUGCAUUACAGUUGCGAGGCAUGCCGUGCGGUGGAGAAAGACAUGGUAUCUUUUAUGAGGCAGACG ---.((((((((((((((.......(((((((.((....))...))))))).........((((((..((((((((...(((((.......))))).)))))))..)..))))))..)))).)))).)).((((((.......))))))....)))).... ( -61.00, z-score = -2.38, R) >droPer1.super_43 338244 158 + 623856 ---AUGUCCCCUGCUGGCUCUGCGACAGCGUAGUGCACGCAAAAUGCGCUGGAUUUUCUGGCCUCGUGAGUGAUGCUAUAGCCAAACGUAAUGGCUUGCAUUACAGUUGCGAGGCAUGCCGUGCGGUGGAGAAAGACAUGGUGGCUUUCAUGAGGCAGACG ---.((((((((((((((.......(((((((.((....))...))))))).........((((((..((((((((...(((((.......))))).)))))))..)..))))))..)))).)))).)).......(((((......))))).)))).... ( -63.90, z-score = -1.88, R) >droWil1.scaffold_181155 1930314 158 - 3442787 ---AUAUUCUUUGUUGGUUGUGUGACAAUGCGGUUCACGUUAAGUGUGCAGGUUACACAGGCAGAAUCAAGGAGUCUAUUGCUAAAGGUGGUGGCUUAAAAUAUGGAUGUGAUGCUUGCCGGGAGGUCGAGAAUGAGAUGCGCUCUUUCAUGAGGCGGACU ---....(((((((((.(((((((((..((((...(((.....))))))).))))))))).)))...))))))((((((..(.....)..)).((.........(.((.(.((.((((((....)).)))).)).).)).).(((......))))))))). ( -44.50, z-score = -0.16, R) >droVir3.scaffold_12736 420639 145 - 612119 AAAUUGUUAAUUGCUGGCUGUAUGAGAAUAUGAUGCAUGCUUAAUGUGUAGGUCUCACUGAGCGUGAUUGCGACAAGAUCGCU----GCAAUUGCCAAGA--AGGGCUGCGGCGGCUUGCGU-UGGUCGCGCCCAGAAUGAAUCGACAAACG--------- ...((((..(((.((((.....(((((.....((((((.....))))))...)))))....(((((((((((.((((.(((((----(((...(((....--..))))))))))))))))).-)))))))))))))....)))..))))...--------- ( -52.90, z-score = -2.39, R) >consensus ___AUGUUCCUUGCUGGCUCUGUGACAGCGUAGUGCACGCAAAAUGCGCAGGAUUUUCUGGCCGCGUGAGUGAUGCUAUAGCCAAACGUAAUGGCUUAAAUUACAGUUGCGAGGCAUGCCGUGCGGUCGAGAAAGACAUGAAAUCUUUCAUGAGGCAGACG ................(((((((....)))))))..........................((((((((((.((((((((.((.....)).)))))......................(((....)))...............))).))))))))))..... (-12.96 = -14.40 + 1.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:00:04 2011