| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,091,978 – 2,092,107 |
| Length | 129 |
| Max. P | 0.957450 |

| Location | 2,091,978 – 2,092,107 |
|---|---|
| Length | 129 |
| Sequences | 7 |
| Columns | 143 |
| Reading direction | reverse |
| Mean pairwise identity | 75.89 |
| Shannon entropy | 0.42538 |
| G+C content | 0.45111 |
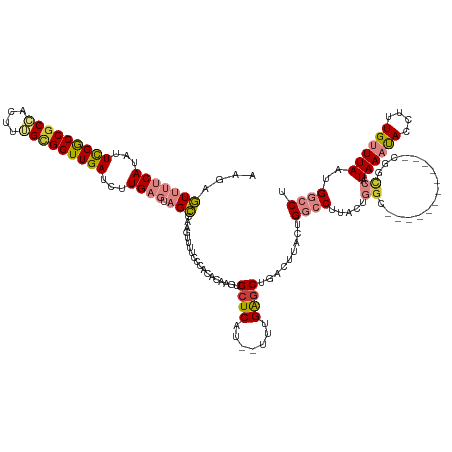
| Mean single sequence MFE | -39.29 |
| Consensus MFE | -20.36 |
| Energy contribution | -21.43 |
| Covariance contribution | 1.07 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.957450 |
| Prediction | RNA |
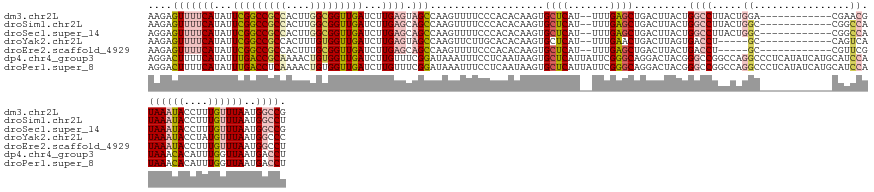
Download alignment: ClustalW | MAF
>dm3.chr2L 2091978 129 - 23011544 AAGAGUUUUCAUAUUCGGCCGCCACUUGGCGGUUGAUCUUGAGUAGCCAAGUUUUCCCACACAAGUGCUCAU--UUUGAGCUGACUUACUGGCCUUACUGGA------------CGAACGUAAAUACCUUUGUUUAAUGGCCG ..(((((((((...(((((((((....)))))))))...)))).(((((((......(((....))).....--)))).))))))))...((((....((((------------((((.((....)).))))))))..)))). ( -40.80, z-score = -3.03, R) >droSim1.chr2L 2062933 129 - 22036055 AAGAGUUUUCAUAUUCGGCCGCCACUUGGCGGUUGAUCUUGAGCAGCCAAGUUUUCCCACACAAGUGCUCAU--UUUGAGCUGACUUACUGGCCUUACUGGC------------CGGCCAUAAAUACCUUUGUUUAAUGGCCU ..((((.((((...(((((((((....)))))))))...))))((((((((......(((....))).....--)))).))))))))...((((.....)))------------)(((((((((((....))))).)))))). ( -48.40, z-score = -4.52, R) >droSec1.super_14 2038277 129 - 2068291 AGGAGUUUUCAUAUUCGGCCGCCACUUGGCGGUUGAUCUUGAGCAGCCAAGUUUUCCCACACAAGUGCUCAU--UUUGAGCUGACUUACUGGCCUUACUGGC------------CGGCCAUAAAUACCUUUGUUUAAUGGCCG ..((((.((((...(((((((((....)))))))))...))))((((((((......(((....))).....--)))).))))))))...((((.....)))------------)(((((((((((....))))).)))))). ( -49.00, z-score = -4.43, R) >droYak2.chr2L 2079574 124 - 22324452 AAGAGUUUUCAUAUUCGGCCGCCACUUUGUGGUUGAUCUUGAGUAGCCAAGUUCUUGCACACAAGUGCUCAU--UUUGAACUGACUUAGUGACCU-----GC------------CAGUCAUAAAUACCUAUGUUUAAUGGCCC .......((((...((((((((......))))))))...))))..(((((((((..((((....))))....--...)))))......(((((..-----..------------..)))))................)))).. ( -31.00, z-score = -1.26, R) >droEre2.scaffold_4929 2143056 124 - 26641161 AAGAGUUUUCAUAUUCGGCCGCCACUUUGCGGUUGAUCUUGAGCAGCCAAGUUUUCCCACACAAGUGCUCAU--UUUGAGCUGACUUACUGACCU-----GC------------CGUUCGUAAAUACCUUUGUUUAAUGGCCU ..((((.((((...((((((((......))))))))...))))((((((((......(((....))).....--)))).))))))))........-----((------------(((...((((((....))))))))))).. ( -31.40, z-score = -1.48, R) >dp4.chr4_group3 8903731 143 - 11692001 AGGACUUUUCAUAUUUGACCGCAAAACUGUGGUUGAUCUUGUUUCGGAUAAAUUUCCUCAAUAAGUGCUCAUUAUUCGGGCAGGACUACGGGCCGGCCAGGCCCUCAUAUCAUGCAUCCAUAAACACAUUUGGUUAAUGACCU ..............(..((((((....))))))..).((((((..(((......)))..))))))(((((.......)))))((.....(((((.....))))).....((((....(((..........)))...)))))). ( -38.50, z-score = -1.28, R) >droPer1.super_8 88093 143 - 3966273 AGGACUUUUCAUAUUUGACCUCAAAACUGUGGUUGAUCUUGUUUCGGAUAAAUUUCCUCAAUAAGUGCUCAUUAUUCGGGCAGGACUACGGGCCGGCCAGGCCCUCAUAUCAUGCAUCCAUAAACACAUUUGGUUAAUGACCU (((............(((..........((((((...((((((..(((......)))..))))))(((((.......))))).))))))(((((.....))))))))..((((....(((..........)))...))))))) ( -35.90, z-score = -0.75, R) >consensus AAGAGUUUUCAUAUUCGGCCGCCACUUUGCGGUUGAUCUUGAGUAGCCAAGUUUUCCCACACAAGUGCUCAU__UUUGAGCUGACUUACUGGCCUUACUGGC____________CGGCCAUAAAUACCUUUGUUUAAUGGCCU ........(((...(((((((((....)))))))))...)))...((((.................(((((.....))))).......................................((((((....)))))).)))).. (-20.36 = -21.43 + 1.07)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:10:10 2011