
| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 1,663,235 – 1,663,294 |
| Length | 59 |
| Max. P | 0.711203 |

| Location | 1,663,235 – 1,663,294 |
|---|---|
| Length | 59 |
| Sequences | 5 |
| Columns | 65 |
| Reading direction | reverse |
| Mean pairwise identity | 57.91 |
| Shannon entropy | 0.73866 |
| G+C content | 0.43030 |
| Mean single sequence MFE | -14.34 |
| Consensus MFE | -6.26 |
| Energy contribution | -5.78 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.91 |
| Mean z-score | -0.74 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.711203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2RHet 1663235 59 - 3288761 -CAGCCAAAA-AUUACUU---GUUGGCUAUGUGAUAGAAUAGUGCAUACCAUUAUGGCCGUACA- -.((((((.(-(....))---.)))))).((((.....((((((.....)))))).....))))- ( -11.20, z-score = -0.03, R) >dp4.chrXL_group1a 8426800 62 + 9151740 -CGGCAAACA-UUCACCUUAUGUUGGCUGUGUGAUAACAUGGCACACACUGUUUGCGCCGACUU- -((((.((((-(.......))))).(((((((....))))))).............))))....- ( -17.70, z-score = -1.39, R) >droPer1.super_26 516902 62 + 1349181 -CGGCAAACA-UUCACCUUCUGUUGGCUGUGUGAUAACAUGGCACACACUGUUUGCGCCGACUU- -..(((((((-..((.(....).))(((((((....)))))))......)))))))........- ( -16.10, z-score = -0.83, R) >droWil1.scaffold_181148 2156648 63 - 5435427 -CAGUGAAGACUUUCUUUACUAUUGGCUGUGUGACAAUGUAAUGCACUUUAAAUGUGCUGGUUA- -.(((((((.....)))))))((((.........)))).....((((.......))))......- ( -12.10, z-score = -0.46, R) >droVir3.scaffold_12736 191424 63 - 612119 CUGCCGCAAA--UUGUUAAUUGCUGGCUGUAUGAGAAUAUGAUGCAUGCUUAAUGUGUAGGUCUC ..(((((((.--.......)))).))).....((((.....((((((.....))))))...)))) ( -14.60, z-score = -0.99, R) >consensus _CGGCAAAAA_UUUACUUACUGUUGGCUGUGUGAUAACAUGGUGCACACUAUUUGCGCCGACUU_ .....................((((((.(((((...........))))).......))))))... ( -6.26 = -5.78 + -0.48)

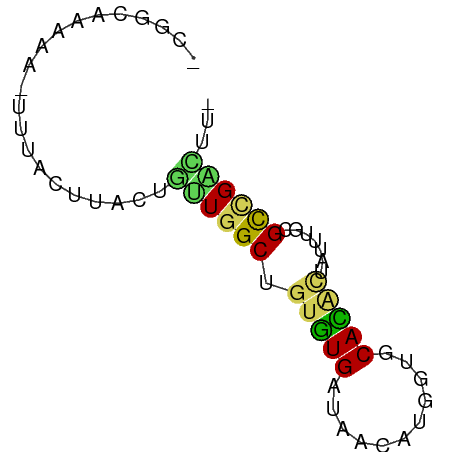
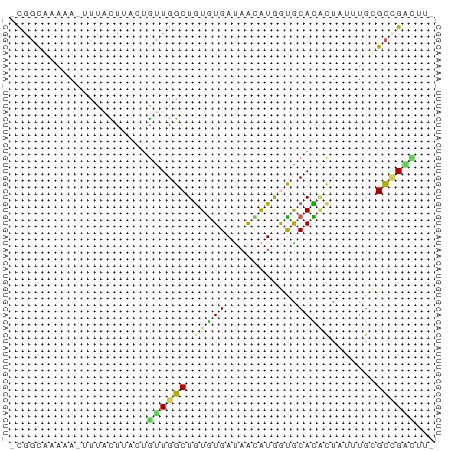
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:59:55 2011