| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 1,649,658 – 1,649,740 |
| Length | 82 |
| Max. P | 0.958619 |

| Location | 1,649,658 – 1,649,740 |
|---|---|
| Length | 82 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 80.52 |
| Shannon entropy | 0.33028 |
| G+C content | 0.47420 |
| Mean single sequence MFE | -26.65 |
| Consensus MFE | -22.89 |
| Energy contribution | -22.58 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.958619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
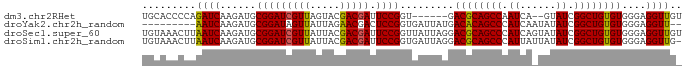
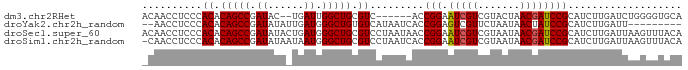
>dm3.chr2RHet 1649658 82 + 3288761 UGCACCCCAGAUCAAGAUGCGGAUCGUUAGUACGACGAUUCCGGU------GACGCAGCCAAUCA--GUAUCGGCUGUGUGGGAGGUUGU .((..((((..(((...((.((((((((.....)))))))))).)------))(((((((.....--.....)))))))))))..))... ( -29.50, z-score = -1.68, R) >droYak2.chr2h_random 471774 79 - 3774259 ---------AAUCAAGAUGCGGAUAGUUAUUAGAACGACUCCGGUGAUUAUGACACAGCCCAUCAAUAUAUCGGCUGUGUGGGAGGUU-- ---------(((((.....((((..(((.....)))...)))).))))).(.((((((((.((......)).)))))))).)......-- ( -20.40, z-score = -0.90, R) >droSec1.super_60 50372 90 + 163194 UGUAAACUUAAUCAAGAUGCGGAUCGUUAUUACGACGAUUCCGGUUAUUAGGACGCAGCCCAUCAGUAUAUCGGCUGUGUGGGAGGUUGU ........(((((....((((.((((((.....))))))(((........))))))).(((((((((......)))).))))).))))). ( -25.70, z-score = -1.15, R) >droSim1.chr2h_random 451525 89 - 3178526 UGUAAACUUAAUCAAGAUGCGGAUCGUUAUUACGACGAUUCCGGUGAUUAGGACGCAGCCCAUUAUUAUAUCGGCUGUGUGGGAGGUUG- ......((((((((...((.((((((((.....)))))))))).))))))))((((((((.((......)).)))))))).........- ( -31.00, z-score = -3.26, R) >consensus UGUAAACUUAAUCAAGAUGCGGAUCGUUAUUACGACGAUUCCGGUGAUUAGGACGCAGCCCAUCA_UAUAUCGGCUGUGUGGGAGGUUG_ .........((((......(((((((((.....))))).)))).........((((((((.((......)).))))))))....)))).. (-22.89 = -22.58 + -0.31)
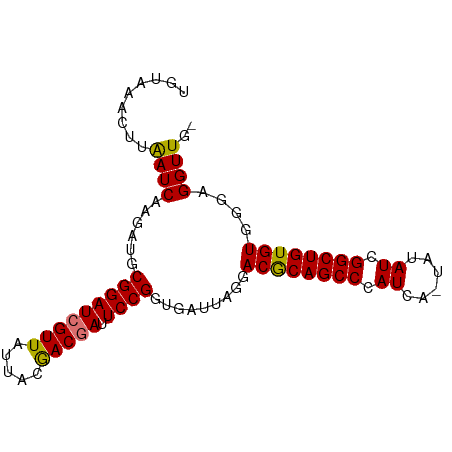
| Location | 1,649,658 – 1,649,740 |
|---|---|
| Length | 82 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 80.52 |
| Shannon entropy | 0.33028 |
| G+C content | 0.47420 |
| Mean single sequence MFE | -21.35 |
| Consensus MFE | -15.96 |
| Energy contribution | -16.02 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.887545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
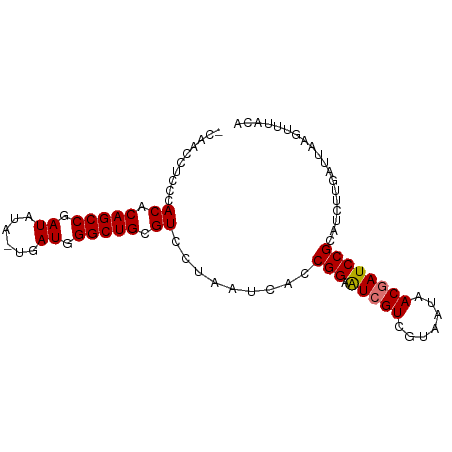
>dm3.chr2RHet 1649658 82 - 3288761 ACAACCUCCCACACAGCCGAUAC--UGAUUGGCUGCGUC------ACCGGAAUCGUCGUACUAACGAUCCGCAUCUUGAUCUGGGGUGCA ..........((.((((((((..--..)))))))).)).------...(((.((((.......)))))))(((((((.....))))))). ( -26.60, z-score = -1.95, R) >droYak2.chr2h_random 471774 79 + 3774259 --AACCUCCCACACAGCCGAUAUAUUGAUGGGCUGUGUCAUAAUCACCGGAGUCGUUCUAAUAACUAUCCGCAUCUUGAUU--------- --........((((((((.((......)).))))))))...(((((.((((...(((.....)))..)))).....)))))--------- ( -19.80, z-score = -2.12, R) >droSec1.super_60 50372 90 - 163194 ACAACCUCCCACACAGCCGAUAUACUGAUGGGCUGCGUCCUAAUAACCGGAAUCGUCGUAAUAACGAUCCGCAUCUUGAUUAAGUUUACA .(((......((.(((((.((......)).))))).)).........((((.((((.......))))))))....)))............ ( -18.20, z-score = -0.76, R) >droSim1.chr2h_random 451525 89 + 3178526 -CAACCUCCCACACAGCCGAUAUAAUAAUGGGCUGCGUCCUAAUCACCGGAAUCGUCGUAAUAACGAUCCGCAUCUUGAUUAAGUUUACA -.........((.(((((.((......)).))))).))..((((((.((((.((((.......)))))))).....))))))........ ( -20.80, z-score = -2.19, R) >consensus _CAACCUCCCACACAGCCGAUAUA_UGAUGGGCUGCGUCCUAAUCACCGGAAUCGUCGUAAUAACGAUCCGCAUCUUGAUUAAGUUUACA ..........((.(((((.((......)).))))).)).........(((.(((((.......))))))))................... (-15.96 = -16.02 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:59:53 2011