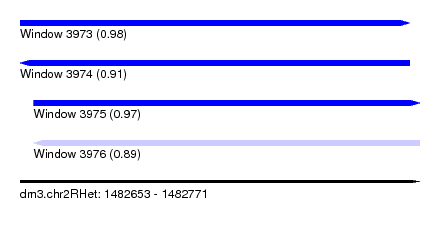
| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 1,482,653 – 1,482,771 |
| Length | 118 |
| Max. P | 0.976414 |

| Location | 1,482,653 – 1,482,768 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 56.81 |
| Shannon entropy | 0.60031 |
| G+C content | 0.48116 |
| Mean single sequence MFE | -33.67 |
| Consensus MFE | -16.59 |
| Energy contribution | -15.60 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.976414 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2RHet 1482653 115 + 3288761 UGAUAGACCAGCACUCGUGCGUGUGGAUAAUGGAACAGCCAUCCACGUCAAAGGAACGCACCUAAUAACAUUCAUUGUGAGUGCCAUGGUCAAUGAGACCGUCUUCACAAACCAC (((.((((..(((((((((((((((((...(((.....)))))))).((....))))))))......(((.....)))))))))...((((.....)))))))))))........ ( -37.50, z-score = -3.10, R) >droEre2.scaffold_4784 2404465 115 - 25762168 CGAUAGAUCAGCACUUGUGCGCGUGGAUAAUGGAACGACCAUCCACAUUAGAGGCACGCACCUGAUAACAUUUACUGAGAGUGCUAUGGUCAAUGAGACCGUAUUUGUAAAUCAU .(((.....(((((((...(..(((((...(((.....))))))))(((((..(....)..)))))..........).)))))))((((((.....))))))........))).. ( -30.50, z-score = -1.34, R) >dp4.Unknown_group_128 8950 115 + 26480 CGCCGCACAUGCCUUCGUAACCAUUGACAAUGGCCCGGAACGCCGAAUUACGGGAAAGUACCUGGUGAAUUUUGACCAGGUAGCCUGCAUGAAGGGUUCUACGAACACCAACUUC .......(((((...(((((((((.....))))..(((....)))..)))))((....((((((((........)))))))).)).)))))..((((((...)))).))...... ( -33.00, z-score = -0.97, R) >consensus CGAUAGACCAGCACUCGUGCGCGUGGAUAAUGGAACGGCCAUCCACAUUAAAGGAACGCACCUGAUAACAUUUAAUGAGAGUGCCAUGGUCAAUGAGACCGUAUUCACAAACCAC .....(((((((((((..............((((.......))))......(((......)))...............))))))..)))))........................ (-16.59 = -15.60 + -0.99)

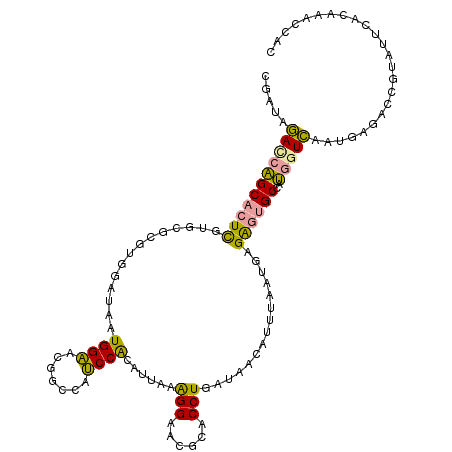
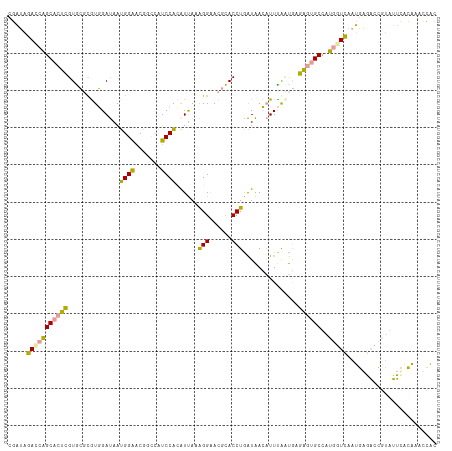
| Location | 1,482,653 – 1,482,768 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 56.81 |
| Shannon entropy | 0.60031 |
| G+C content | 0.48116 |
| Mean single sequence MFE | -35.50 |
| Consensus MFE | -16.85 |
| Energy contribution | -15.76 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2RHet 1482653 115 - 3288761 GUGGUUUGUGAAGACGGUCUCAUUGACCAUGGCACUCACAAUGAAUGUUAUUAGGUGCGUUCCUUUGACGUGGAUGGCUGUUCCAUUAUCCACACGCACGAGUGCUGGUCUAUCA ........(((((((((((.....))))..(((((((.(((.((((((........))))))..)))..((((((((........))))))))......))))))).)))).))) ( -38.90, z-score = -1.75, R) >droEre2.scaffold_4784 2404465 115 + 25762168 AUGAUUUACAAAUACGGUCUCAUUGACCAUAGCACUCUCAGUAAAUGUUAUCAGGUGCGUGCCUCUAAUGUGGAUGGUCGUUCCAUUAUCCACGCGCACAAGUGCUGAUCUAUCG ..(((..........((((.....)))).(((((((....(((..((....))..)))((((......((((((((((......)))))))))).)))).)))))))....))). ( -33.00, z-score = -2.25, R) >dp4.Unknown_group_128 8950 115 - 26480 GAAGUUGGUGUUCGUAGAACCCUUCAUGCAGGCUACCUGGUCAAAAUUCACCAGGUACUUUCCCGUAAUUCGGCGUUCCGGGCCAUUGUCAAUGGUUACGAAGGCAUGUGCGGCG ((((..(((.........)))))))....(((.((((((((........)))))))))))..(((((.....((.(((..((((((.....))))))..))).))...))))).. ( -34.60, z-score = -0.46, R) >consensus GUGGUUUGUGAACACGGUCUCAUUGACCAUGGCACUCUCAAUAAAUGUUAUCAGGUGCGUUCCUCUAAUGUGGAUGGCCGUUCCAUUAUCCACGCGCACGAGUGCUGGUCUAUCG .....................((.(((((..((((((.((.....)).......((((((.........(((((.......))))).......))))))))))))))))).)).. (-16.85 = -15.76 + -1.10)

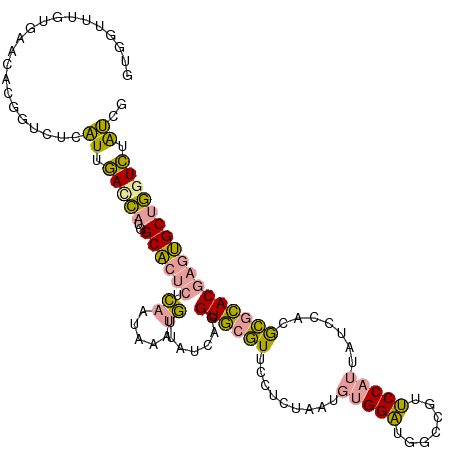
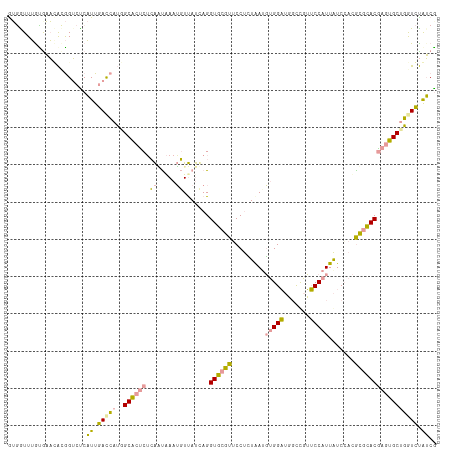
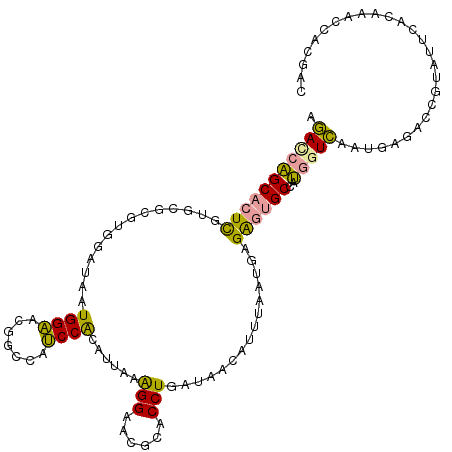
| Location | 1,482,657 – 1,482,771 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 57.02 |
| Shannon entropy | 0.59752 |
| G+C content | 0.47661 |
| Mean single sequence MFE | -32.77 |
| Consensus MFE | -16.59 |
| Energy contribution | -15.60 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.974215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2RHet 1482657 114 + 3288761 AGACCAGCACUCGUGCGUGUGGAUAAUGGAACAGCCAUCCACGUCAAAGGAACGCACCUAAUAACAUUCAUUGUGAGUGCCAUGGUCAAUGAGACCGUCUUCACAAACCACGAC ((((..(((((((((((((((((...(((.....)))))))).((....))))))))......(((.....)))))))))...((((.....)))))))).............. ( -35.50, z-score = -2.41, R) >droEre2.scaffold_4784 2404469 114 - 25762168 AGAUCAGCACUUGUGCGCGUGGAUAAUGGAACGACCAUCCACAUUAGAGGCACGCACCUGAUAACAUUUACUGAGAGUGCUAUGGUCAAUGAGACCGUAUUUGUAAAUCAUGAC .(((.(((((((...(..(((((...(((.....))))))))(((((..(....)..)))))..........).)))))))((((((.....))))))........)))..... ( -29.80, z-score = -1.16, R) >dp4.Unknown_group_128 8954 114 + 26480 GCACAUGCCUUCGUAACCAUUGACAAUGGCCCGGAACGCCGAAUUACGGGAAAGUACCUGGUGAAUUUUGACCAGGUAGCCUGCAUGAAGGGUUCUACGAACACCAACUUCAAU ...(((((...(((((((((.....))))..(((....)))..)))))((....((((((((........)))))))).)).)))))..((((((...)))).))......... ( -33.00, z-score = -1.52, R) >consensus AGACCAGCACUCGUGCGCGUGGAUAAUGGAACGGCCAUCCACAUUAAAGGAACGCACCUGAUAACAUUUAAUGAGAGUGCCAUGGUCAAUGAGACCGUAUUCACAAACCACGAC .(((((((((((..............((((.......))))......(((......)))...............))))))..)))))........................... (-16.59 = -15.60 + -0.99)

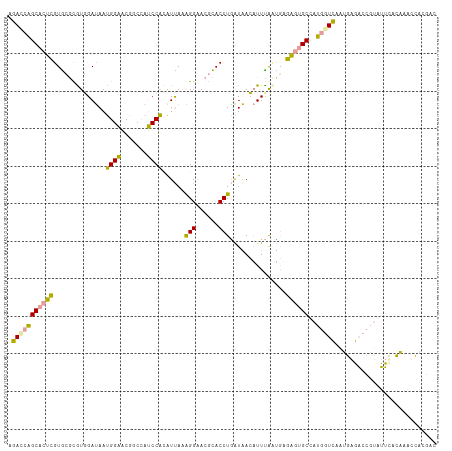
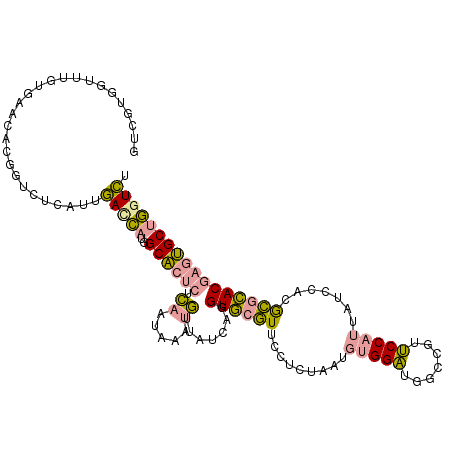
| Location | 1,482,657 – 1,482,771 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 57.02 |
| Shannon entropy | 0.59752 |
| G+C content | 0.47661 |
| Mean single sequence MFE | -34.73 |
| Consensus MFE | -16.66 |
| Energy contribution | -16.22 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.885132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2RHet 1482657 114 - 3288761 GUCGUGGUUUGUGAAGACGGUCUCAUUGACCAUGGCACUCACAAUGAAUGUUAUUAGGUGCGUUCCUUUGACGUGGAUGGCUGUUCCAUUAUCCACACGCACGAGUGCUGGUCU ..(((((((.((((........)))).)))))))((((((.(((.((((((........))))))..)))..((((((((........))))))))......))))))...... ( -38.40, z-score = -1.44, R) >droEre2.scaffold_4784 2404469 114 + 25762168 GUCAUGAUUUACAAAUACGGUCUCAUUGACCAUAGCACUCUCAGUAAAUGUUAUCAGGUGCGUGCCUCUAAUGUGGAUGGUCGUUCCAUUAUCCACGCGCACAAGUGCUGAUCU .....(((..........((((.....)))).(((((((....(((..((....))..)))((((......((((((((((......)))))))))).)))).)))))))))). ( -32.70, z-score = -2.05, R) >dp4.Unknown_group_128 8954 114 - 26480 AUUGAAGUUGGUGUUCGUAGAACCCUUCAUGCAGGCUACCUGGUCAAAAUUCACCAGGUACUUUCCCGUAAUUCGGCGUUCCGGGCCAUUGUCAAUGGUUACGAAGGCAUGUGC ..........((.((((((((((((....(((.((.((((((((........))))))))....)).)))....)).))))...(((((.....))))))))))).))...... ( -33.10, z-score = -0.70, R) >consensus GUCGUGGUUUGUGAACACGGUCUCAUUGACCAUGGCACUCUCAAUAAAUGUUAUCAGGUGCGUUCCUCUAAUGUGGAUGGCCGUUCCAUUAUCCACGCGCACGAGUGCUGGUCU ...........................(((((..((((((.((.....)).......((((((.........(((((.......))))).......))))))))))))))))). (-16.66 = -16.22 + -0.43)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:59:50 2011