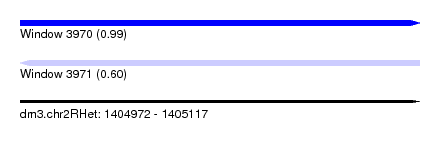
| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 1,404,972 – 1,405,117 |
| Length | 145 |
| Max. P | 0.986113 |

| Location | 1,404,972 – 1,405,117 |
|---|---|
| Length | 145 |
| Sequences | 4 |
| Columns | 145 |
| Reading direction | forward |
| Mean pairwise identity | 94.48 |
| Shannon entropy | 0.08952 |
| G+C content | 0.49138 |
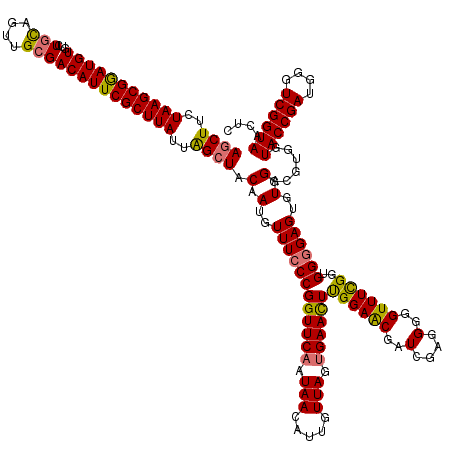
| Mean single sequence MFE | -35.80 |
| Consensus MFE | -29.21 |
| Energy contribution | -30.02 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.02 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.986113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
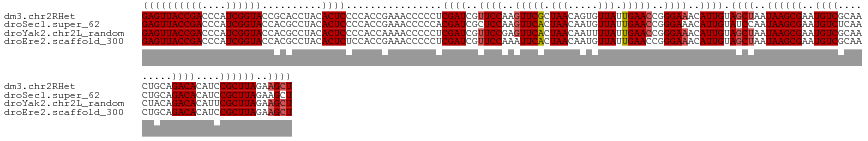
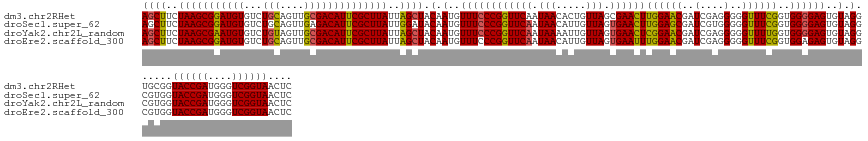
>dm3.chr2RHet 1404972 145 + 3288761 GAGUUACCGACCCAUCGGUACCGCACCUACACUCCCCACCGAAACCCCCUCGAUCGUUCCAAGUUCGCUAACAGUGUUAUUGAACCGGGAAACAUUGUAGCUAAUAAGCGAAUGUCGCAACUGCAGACACAUCCGCUUAGAAGCU ((((((((((....))))))..........))))................((((..((((..(((((.((((...)))).)))))..))))..)))).((((..((((((..((((((....)).))))....))))))..)))) ( -37.10, z-score = -2.51, R) >droSec1.super_62 3693 145 + 174595 GAGUUACCGACCCAUCGGUACCACGCCUACACUCCCCACCGAAACCCCCACGAUCGCUCCAAGUUCACUAACAAUGUUAUUGAACCGGGAAACAUUGUAUCCAAUAAGCGAAUGUCUCAACUGCAGACACAUCCGCUUAGAAGCU ..(.((((((....)))))).)..((.......................(((((...(((..(((((.((((...)))).)))))..)))...)))))......((((((.((((.((.......)).)))).))))))...)). ( -32.70, z-score = -2.77, R) >droYak2.chr2L_random 1337233 145 + 4064425 GAGUUACCGACCCAUCGGUACCACGCCUACACUCCCCACCAAAACCCCCUCGAUCGUUCCGAGUUCACUAACAAUUUUAUUGAACCGGGAAACAUUGUAGCUAAUAAGCGAAUGUCGCAACUACAGACACAUUCGCUUAGAAGCU ((((((((((....))))))..........))))................((((..(((((.(((((.(((.....))).))))))))))...)))).((((..(((((((((((..(.......)..)))))))))))..)))) ( -38.80, z-score = -4.82, R) >droEre2.scaffold_300 1233 145 + 4206 GAGUUACCGACCCAUCGGUACCACGCCUACACUCUCCACCGAAACCCCCUCGAUCGUUCCAAAUUCACUAACAAUGUUAUUGAACCGGGAAACAUUGUAGCUAAUAAGCGAAUGUCGCAACUGCAGACACAUCCGCUUAGAAGCU ((((((((((....))))))..........))))................((((..((((...((((.((((...)))).))))...))))..)))).((((..((((((..((((((....)).))))....))))))..)))) ( -34.60, z-score = -2.82, R) >consensus GAGUUACCGACCCAUCGGUACCACGCCUACACUCCCCACCGAAACCCCCUCGAUCGUUCCAAGUUCACUAACAAUGUUAUUGAACCGGGAAACAUUGUAGCUAAUAAGCGAAUGUCGCAACUGCAGACACAUCCGCUUAGAAGCU ((((((((((....))))))..........))))................((((..((((..(((((.(((.....))).)))))..))))..)))).((((..((((((..((((.........))))....))))))..)))) (-29.21 = -30.02 + 0.81)
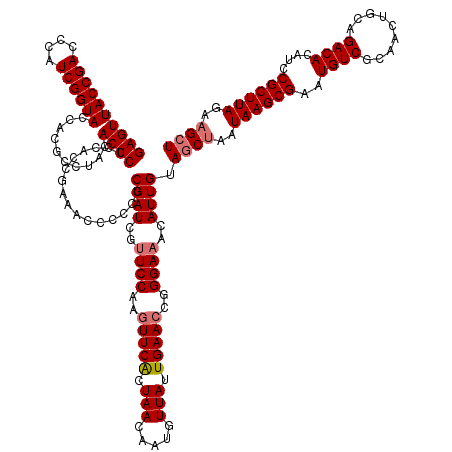
| Location | 1,404,972 – 1,405,117 |
|---|---|
| Length | 145 |
| Sequences | 4 |
| Columns | 145 |
| Reading direction | reverse |
| Mean pairwise identity | 94.48 |
| Shannon entropy | 0.08952 |
| G+C content | 0.49138 |
| Mean single sequence MFE | -46.17 |
| Consensus MFE | -42.36 |
| Energy contribution | -42.05 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.600749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2RHet 1404972 145 - 3288761 AGCUUCUAAGCGGAUGUGUCUGCAGUUGCGACAUUCGCUUAUUAGCUACAAUGUUUCCCGGUUCAAUAACACUGUUAGCGAACUUGGAACGAUCGAGGGGGUUUCGGUGGGGAGUGUAGGUGCGGUACCGAUGGGUCGGUAACUC ((((..(((((((((((...(((....))))))))))))))..)))).(.(..(((((((((((..((((...))))..)))))((((((..(....)..))))))..))))))..).)......((((((....)))))).... ( -46.80, z-score = -0.84, R) >droSec1.super_62 3693 145 - 174595 AGCUUCUAAGCGGAUGUGUCUGCAGUUGAGACAUUCGCUUAUUGGAUACAAUGUUUCCCGGUUCAAUAACAUUGUUAGUGAACUUGGAGCGAUCGUGGGGGUUUCGGUGGGGAGUGUAGGCGUGGUACCGAUGGGUCGGUAACUC .((((.(((((((((((.((.......)).))))))))))).........(..((((((((((((.((((...)))).))))))((((((..(....)..))))))..))))))..)))))....((((((....)))))).... ( -47.40, z-score = -1.67, R) >droYak2.chr2L_random 1337233 145 - 4064425 AGCUUCUAAGCGAAUGUGUCUGUAGUUGCGACAUUCGCUUAUUAGCUACAAUGUUUCCCGGUUCAAUAAAAUUGUUAGUGAACUCGGAACGAUCGAGGGGGUUUUGGUGGGGAGUGUAGGCGUGGUACCGAUGGGUCGGUAACUC ((((..(((((((((((((........)).)))))))))))..)))).(.(..((((((((((((.(((.....))).))))))((((((..(....)..))))))..))))))..).)......((((((....)))))).... ( -45.30, z-score = -1.58, R) >droEre2.scaffold_300 1233 145 - 4206 AGCUUCUAAGCGGAUGUGUCUGCAGUUGCGACAUUCGCUUAUUAGCUACAAUGUUUCCCGGUUCAAUAACAUUGUUAGUGAAUUUGGAACGAUCGAGGGGGUUUCGGUGGAGAGUGUAGGCGUGGUACCGAUGGGUCGGUAACUC ((((..(((((((((((...(((....))))))))))))))..))))...((((((((..(((((.((((...)))).)))))..))))(.((((((.....)))))).).........))))..((((((....)))))).... ( -45.20, z-score = -1.19, R) >consensus AGCUUCUAAGCGGAUGUGUCUGCAGUUGCGACAUUCGCUUAUUAGCUACAAUGUUUCCCGGUUCAAUAACAUUGUUAGUGAACUUGGAACGAUCGAGGGGGUUUCGGUGGGGAGUGUAGGCGUGGUACCGAUGGGUCGGUAACUC ((((..(((((((((((...(((....))))))))))))))..)))).(.(..((((((((((((.(((.....))).))))))((((((..(....)..))))))..))))))..).)......((((((....)))))).... (-42.36 = -42.05 + -0.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:59:46 2011