| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 1,139,417 – 1,139,512 |
| Length | 95 |
| Max. P | 0.932800 |

| Location | 1,139,417 – 1,139,512 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 50.81 |
| Shannon entropy | 1.02508 |
| G+C content | 0.33134 |
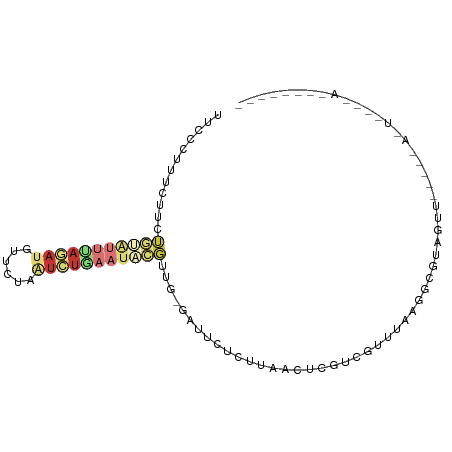
| Mean single sequence MFE | -17.08 |
| Consensus MFE | -3.56 |
| Energy contribution | -5.01 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.73 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.21 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932800 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
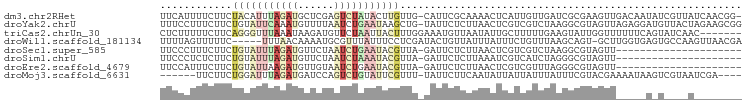
>dm3.chr2RHet 1139417 95 - 3288761 UUCAUUUUCUUCUACAUUUAGAUGCUCGAGUCUAUACUUGUUG-CAUUCGCAAAACUCAUUGUUGAUCGCGAAGUUGACAAUAUCGUUAUCAACGG- ....................(((((.(((((....)))))..)-))))............(((((((.((((.((.....)).)))).))))))).- ( -18.60, z-score = -1.00, R) >droYak2.chrU 10295500 96 + 28119190 UUUCCUUUCUUCUGUAUUCAAAUGUUUUAAUCUGAAUAAGCUG-UAUUCUCUUAACUCGUCGUCUAAGGCGUAGUUAGAGGAUGUUACUAGAAGCGG ...((...((((((((((((.((......)).)))))).....-((((((((.((((((((......)))).))))))))))))....)))))).)) ( -24.80, z-score = -2.79, R) >triCas2.chrUn_30 95370 90 - 281566 CUCUUUUUCUUCAGGGUUUAAAUAAGAUGUUCUAAUUACUUUGGAAAUGUUAAUAUUGCUUUUUGAAGUAUUGGUUUUUUCAGUAUCAAC------- ........(((((((((.....(((.((.((((((.....)))))))).))).....)))..))))))((((((.....)))))).....------- ( -12.70, z-score = -0.23, R) >droWil1.scaffold_181134 257337 91 - 1305803 UUUUAGUUUUUC-----UUUAACAAAAUGCGUUUAUUUCCUCGAUACUGUUAUUUAUUUCUGUUUAAGCAGU-GCUUGGUGAGUGCCAAGUUAACGA ............-----............((((............((((((...............))))))-(((((((....))))))).)))). ( -13.76, z-score = -0.00, R) >droSec1.super_585 9071 75 + 12446 UUCCCUUUCUUCUGUAUUUAGAUGUUCUAAUCUGAAUACGUUA-GAUUCUCUUAACUCGUCGUCUAAGGCGUAGUU--------------------- .......(((..(((((((((((......)))))))))))..)-)).......((((((((......)))).))))--------------------- ( -20.70, z-score = -4.61, R) >droSim1.chrU 12889202 75 + 15797150 UUCCCUCUCUUCUGUAUUUAGAUGUUCUAAUCUAAAUACGUUA-GAUUCUCUUAAAUCGUCAUCUAGGGCGUAGUU--------------------- .......(((..(((((((((((......)))))))))))..)-))...........((((......)))).....--------------------- ( -16.90, z-score = -3.68, R) >droEre2.scaffold_4679 6871 75 + 42690 UUCCAUUUCUUCUGUAUUAAGAUGUUGUAAUCUGAAUACGUUA-GAUUCUCUUAACUCGUCGUUUAGGGCGUAGUU--------------------- .......(((..((((((.((((......)))).))))))..)-)).......((((((((......)))).))))--------------------- ( -16.10, z-score = -2.12, R) >droMoj3.scaffold_6631 99 86 - 1979 ------UUCUUCUGGAUUUAGAUGAUCCAGUCUGUAUUCGUUU-UAUUCUUCAAUAUUAUUAUUUAUUUCGUACGAAAAUAAGUCGUAAUCGA---- ------.....(((((((.....))))))).............-...........(((((.(((((((((....).)))))))).)))))...---- ( -13.10, z-score = -0.45, R) >consensus UUCCCUUUCUUCUGUAUUUAGAUGUUCUAAUCUGAAUACGUUG_GAUUCUCUUAACUCGUCGUUUAAGGCGUAGUU_____A_U____A________ ............(((((((((((......)))))))))))......................................................... ( -3.56 = -5.01 + 1.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:59:37 2011