| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 1,123,093 – 1,123,205 |
| Length | 112 |
| Max. P | 0.544736 |

| Location | 1,123,093 – 1,123,205 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 54.12 |
| Shannon entropy | 0.65743 |
| G+C content | 0.44758 |
| Mean single sequence MFE | -31.64 |
| Consensus MFE | -11.69 |
| Energy contribution | -9.15 |
| Covariance contribution | -2.54 |
| Combinations/Pair | 1.72 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.544736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
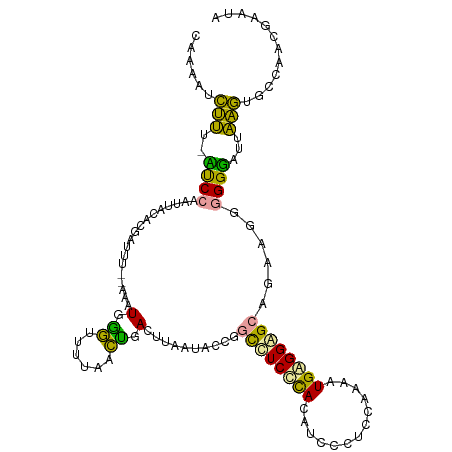
>dm3.chr2RHet 1123093 112 - 3288761 UAAAGCCUUUGAUCCAAGUAUCCGUUUC--AUAUGGAGUUUAAUUGAGCGAACUCCAGCUUCCCACAUCCCUCCAAAAUGGGGAGCAGACGGGGGUAUAAAGUGCCAAGAAAUC ....(((((((((((......((((((.--...((((((((........))))))))((((((((.............)))))))))))))))))).))))).))......... ( -35.02, z-score = -2.59, R) >apiMel3.GroupUn 265390425 111 + 399230636 CAAUAUCGUU-UCCCGAUUACUCGCUUU--AGAUGAGUUUUUGCUGACUUAAUAGAGCUCUCUUACAAAUUCCCAAAAUGAGGUGUACGAGGAGGGAUUAUGUGCCAUUGUAUU ((((..(((.-((((........(((((--(..((((((......)))))).))))))((((.((((..((........))..)))).)))).))))..)))....)))).... ( -25.40, z-score = -0.93, R) >triCas2.chrUn_298 5936 114 + 21235 GAGGAUUUGCUCUCCUAUAAGACGAUGUAAAAAUGGGUUUUGACCGACUUAAUGCCGGCCUCCCAAAUCCCUCCAAAGUGAGGGGCUGAUGGGGGGUUUCAAUGCCACCGAAGA ..((.(((((((((......)).)).)))))....(((.(((((((.(.....).)))(((((((..((((((......))))))....)))))))..)))).))).))..... ( -34.50, z-score = -0.23, R) >consensus CAAAAUCUUU_AUCCAAUUACACGAUUU__AAAUGGGUUUUAACUGACUUAAUACCGGCCUCCCACAUCCCUCCAAAAUGAGGAGCAGAAGGGGGGAUUAAGUGCCAACGAAUA ......(((..((((..................(.((......)).)..........((((((((.............))))))))......))))...)))............ (-11.69 = -9.15 + -2.54)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:59:33 2011