| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 1,086,810 – 1,086,915 |
| Length | 105 |
| Max. P | 0.543996 |

| Location | 1,086,810 – 1,086,915 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 81.28 |
| Shannon entropy | 0.35715 |
| G+C content | 0.50879 |
| Mean single sequence MFE | -34.08 |
| Consensus MFE | -26.78 |
| Energy contribution | -26.36 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.543996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
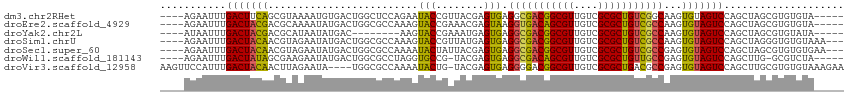
>dm3.chr2RHet 1086810 105 - 3288761 ----AGAAUUUGACUUCAGCGUAAAAUGUGACUGGCUCCAGAAUACCGUUACGAGUGAGGCGACGGCGUUGUCGCGCUGUCGGCAAGUGUAGUCCAGCUAGCGUGUGUA----- ----.........(((((.(((((...(((.(((....)))..)))..)))))..)))))(((((((((....)))))))))(((..(((((.....)).)))..))).----- ( -31.80, z-score = 0.32, R) >droEre2.scaffold_4929 2883978 105 + 26641161 ----AGAAUUUGACUACGACGCAAAAUAUGACUGGCGCCAAAGUACCGAAACGAGUAAGGUGACAGCGUUGUCGCGCUGUCGCCAAGUGUAGUCCAGCUAGCGUGUGUA----- ----..........((((((((.......(((((.(((.....(((((...)).))).(((((((((((....)))))))))))..))))))))......)))).))))----- ( -34.32, z-score = -1.42, R) >droYak2.chr2L 1262169 97 + 22324452 ----AUAAUUUGACUACGACGCAUAAUAUGAC--------AAGUACCGAAAUGAGUGAGGCGACGGCGUUGUCGCGCUGUCGCCAAGUGUAGUCCAGCUAGCGUGUAUA----- ----.......((((((.((............--------...(((........))).(((((((((((....)))))))))))..))))))))...............----- ( -29.80, z-score = -0.87, R) >droSim1.chrU 2817771 107 - 15797150 ----AGAAUUUGACUACAACGUAGAAUAUGACUGGCGCCAAAGUACCGUUAUGAGUGAGGCGACGGCGUUGUCGCGCUGUCGCCAAGUGUAGUCCAGCUAGGGUGUGUAAA--- ----.......(((((((........((((((.((..(....)..)))))))).....(((((((((((....)))))))))))...))))))).................--- ( -37.30, z-score = -1.92, R) >droSec1.super_60 44208 107 - 163194 ----AGAAUUUGACUACAACGUAGAAUAUGACUGGCGCCAAAAUACUAUUACGAGUGAGGCGACGGCGUUGUCGCGCUGUCGCCGAGUGUAGUCCAGCUAGCGUGUGUGAA--- ----..........((((((((((.....(((((.((((....((((......)))).(((((((((((....)))))))))))).))))))))...)).)))).))))..--- ( -37.20, z-score = -1.75, R) >droWil1.scaffold_181143 888035 103 + 1323777 ----AGAAUUUGACUAUAGCGAAGAAUAUGACUGGCGCCUAGGUGCCG-UACGAGUGAGGCGACAGCGUUGUCGCGCUGUUGCCGAGUGUAGUCCAGCUUG-GCGUCUA----- ----.......(((....((.(((.....(((.(((((....))))).-((((.....(((((((((((....)))))))))))...)))))))...))).-)))))..----- ( -40.60, z-score = -2.21, R) >droVir3.scaffold_12958 2137189 109 - 3547706 AAGUUCCAUUUGACUACAACUUAGAAUA----UGGCGCCAAAAUACUG-UACGAGUGAGGGGACGGCGUUGUCGCGCUGACGCCGAGUGUAGUCCAGCUUGCGUGUGUAAAGAA ..((.......(((((((.(((.....(----..(((((....((((.-....)))).(....))))))..).(((....))).))))))))))......))............ ( -27.52, z-score = 1.01, R) >consensus ____AGAAUUUGACUACAACGUAGAAUAUGACUGGCGCCAAAGUACCGUUACGAGUGAGGCGACGGCGUUGUCGCGCUGUCGCCAAGUGUAGUCCAGCUAGCGUGUGUA_____ ...........(((((((.........................(((........))).(((((((((((....)))))))))))...))))))).................... (-26.78 = -26.36 + -0.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:59:32 2011