| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 1,077,320 – 1,077,466 |
| Length | 146 |
| Max. P | 0.959211 |

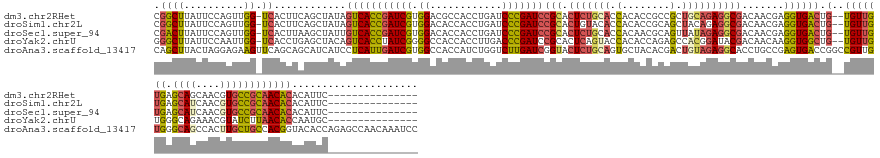
| Location | 1,077,320 – 1,077,466 |
|---|---|
| Length | 146 |
| Sequences | 5 |
| Columns | 164 |
| Reading direction | reverse |
| Mean pairwise identity | 70.69 |
| Shannon entropy | 0.49037 |
| G+C content | 0.55055 |
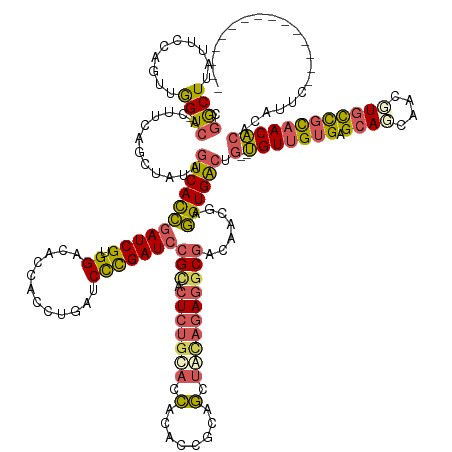
| Mean single sequence MFE | -51.21 |
| Consensus MFE | -29.42 |
| Energy contribution | -30.18 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.959211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2RHet 1077320 146 - 3288761 CGGCUUAUUCCAGUUGG-UCACUUCAGCUAUAGUCACCGAUCGUGGACGCCACCUGAUCCCGAUCCGCACUCUGCACCACACCGCCGCUGCAGAGGCGACAACGAGGUGACUG--UGUUGUGAGCAGCAACGUGCCGCAACACACAUUC--------------- .((......))((((((-.....))))))..((((((((((((.(((((.....)).))))))))(((.(((((((.(........).)))))))))).......)))))))(--(((((((.(((......)))))))))))......--------------- ( -56.80, z-score = -3.11, R) >droSim1.chr2L 950963 146 + 22036055 CGGCUUAUUCCAGUUGG-UCACUUCAGCUAUAGUCACCGAUCGUGGACACCACCUGAUCCCGAUCCGCACUGUACACCACACCGCAGCUACAGAGGCGACAACGAGGUGACUG--UGUUGUGAGCAUCAACGUGCCGCAACACACAUUC--------------- .((......))((((((-.....))))))..((((((((((((.(((((.....)).))))))))(((.(((((..............)))))..))).......)))))))(--(((((((.((((....))))))))))))......--------------- ( -50.04, z-score = -2.47, R) >droSec1.super_94 86510 146 - 93855 CGACUUAUUCCAGUUGG-UCACUUAAGCUAUUGUCACCGAUCGUGGACACCACCUGAUCCCGAUCCGCACUCUGCACCACAACGCAGUUAUAGAGGCGACAACGAGGUGACUG--UGUUGUGAGCAUCAACGUGCCGCAACACACAUUC--------------- (((((......)))))(-((((((......(((((.(((((((.(((((.....)).))))))))......((((........)))).......)).)))))..)))))))((--(((((((.((((....))))))))))))).....--------------- ( -50.20, z-score = -3.08, R) >droYak2.chrU 2676738 146 - 28119190 GGGCUUAUUCCAAUUGG-UCACCUGAGCUACAGUCACCUAUCGGGGCCACCACCUUGACCCGAUCCGCACUCAGUACCACACCAGAGCCACGGAUACGACAACAAGGUGGCUG--UGUUGUGGGCAGAAACGUAUCUUAACACCAAUGC--------------- (((.....))).(((((-(..((..(((.((((((((((.((((((......))))))..(((((((..(((............)))...))))).))......)))))))))--))))..))..(((......)))....))))))..--------------- ( -41.10, z-score = 0.27, R) >droAna3.scaffold_13417 3989137 164 - 6960332 CAGCUUACUAGGAGAAGUUCAGCAGCAUCAUCCUCAUUGAUCGUGGCCACCAUCUGGUCUUGAUCGGUACUCUGCAGUGCUACACGACUGUAGAGGCACCUGCCGAGUGACCGGCCGUUGUGGGCAGCCACUUGCUGCCACGGUACACCAGAGCCAACAAAUCC ..........(((...((((.(((((((((.......)))..(((((..((((((((((....(((((((((((((((........))))))))).....))))))..)))))).....))))...))))).))))))...((....)).)))).......))) ( -57.90, z-score = -0.98, R) >consensus CGGCUUAUUCCAGUUGG_UCACUUCAGCUAUAGUCACCGAUCGUGGACACCACCUGAUCCCGAUCCGCACUCUGCACCACACCGCAGCUACAGAGGCGACAACGAGGUGACUG__UGUUGUGAGCAGCAACGUGCCGCAACACACAUUC_______________ ..............................(((((((((((((.((............)))))))(((.(((((((.(........).)))))))))).......))))))))..(((((((.((((....)))))))))))...................... (-29.42 = -30.18 + 0.76)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:59:30 2011