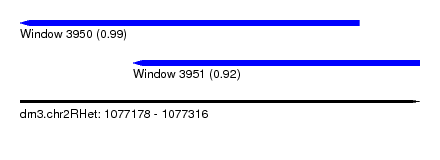
| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 1,077,178 – 1,077,316 |
| Length | 138 |
| Max. P | 0.992933 |

| Location | 1,077,178 – 1,077,295 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.75 |
| Shannon entropy | 0.38798 |
| G+C content | 0.55373 |
| Mean single sequence MFE | -42.13 |
| Consensus MFE | -29.16 |
| Energy contribution | -31.16 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.992933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
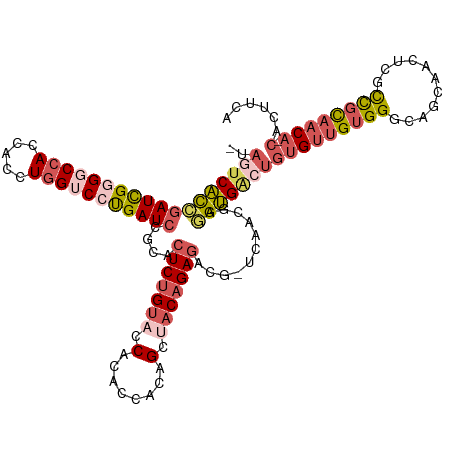
>dm3.chr2RHet 1077178 117 - 3288761 -UAGUCACCGAUCGUGGCCACCACCUGGUCCUGAUCCACACUCUGUACCACACCACAGCUACAGAGGCG-ACAACGAGGUGACUGUGUUGUGGGCAGCACCUCGCCGCAACA-CACUUCA -.((((((((((((.(((((.....))))).)))))....(((((((.(........).)))))))...-.......)))))))(((((((((.(........)))))))))-)...... ( -46.00, z-score = -2.92, R) >droSim1.chrU 15620837 117 + 15797150 -UAUUCACCGAUUGGGGCCACCACUUGGUCCUGAUCCGCACUCUGUACCACACCACAGCAACAGAGACG-UCAACGAGGUGACUGUGUUGUGGGCAGCAACUCGCCGCAACA-CACUUCA -...((((((((..((((((.....))))))..)))....((((((..............))))))...-.......))))).((((((((((.(........)))))))))-))..... ( -43.44, z-score = -2.83, R) >droSec1.super_94 86368 117 - 93855 -UAGUCGCCGAUCGGGGCCACCACCUGGUCCUGAUCUGCACUCUGUACCACACCACAGCUACAGAGACG-UCAACGAAGCGACUGUGUUGUGGGCAGCAACUCGCCGCAACA-CACUUCA -.((((((.(((((((((((.....)))))))))))....(((((((.(........).)))))))...-........))))))(((((((((.(........)))))))))-)...... ( -50.00, z-score = -4.24, R) >droYak2.chrU 2674141 118 - 28119190 -CAGUCACCUAUCGGGGCCACCACCUCGACCCGAUCCGCACUCAGUACCACACCAGAGCCACGGAUACG-ACAACAAGGUGGCUGUGUUGUGGGCAGAAACGUAUCUUAACACCAAUGCA -.((((((((.((((((......))))))..(((((((..(((............)))...))))).))-......))))))))((((((((...(((......)))...)).)))))). ( -35.10, z-score = -0.77, R) >droAna3.scaffold_13417 3988922 116 - 6960332 AUCACCAUUGAUUGUGGCCAUCACCUGGACCCGAUCCGCACUCUGAAUCACUCUACAGUUGCAGAGGCAUUCAACAUAGUGACC--GAUGUGGGCAGCAACGAGCUGU-CCA-AAAUUAA .((((....(((((.(((((.....))).))))))).((.(((((....(((....)))..)))))))..........))))..--....((((((((.....)))))-)))-....... ( -36.10, z-score = -2.12, R) >consensus _UAGUCACCGAUCGGGGCCACCACCUGGUCCUGAUCCGCACUCUGUACCACACCACAGCUACAGAGACG_UCAACGAGGUGACUGUGUUGUGGGCAGCAACUCGCCGCAACA_CACUUCA .(((((((((((((((((((.....)))))))))))....(((((((.(........).)))))))...........)))))))).(((((((...........)))))))......... (-29.16 = -31.16 + 2.00)

| Location | 1,077,217 – 1,077,316 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.97 |
| Shannon entropy | 0.51063 |
| G+C content | 0.54571 |
| Mean single sequence MFE | -26.51 |
| Consensus MFE | -17.16 |
| Energy contribution | -18.36 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.921535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
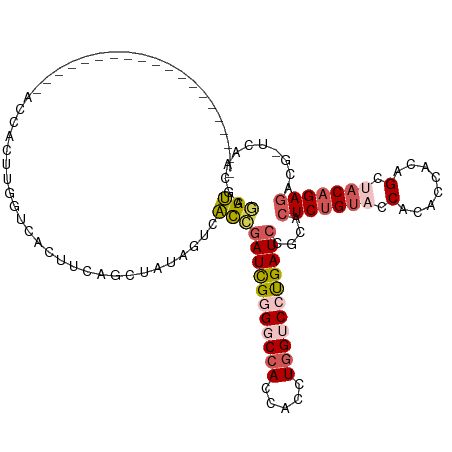
>dm3.chr2RHet 1077217 99 - 3288761 --------------------ACCACUUGGUCACUUCAGCUAUAGUCACCGAUCGUGGCCACCACCUGGUCCUGAUCCACACUCUGUACCACACCACAGCUACAGAGGCG-ACAACGAGGU --------------------(((....(((.(((........))).)))(((((.(((((.....))))).)))))....(((((((.(........).)))))))...-.......))) ( -27.00, z-score = -0.77, R) >droSim1.chrU 15620876 99 + 15797150 --------------------ACCCCUUGAUCACUUCAGCUUUAUUCACCGAUUGGGGCCACCACUUGGUCCUGAUCCGCACUCUGUACCACACCACAGCAACAGAGACG-UCAACGAGGU --------------------...(((((.....................(((..((((((.....))))))..)))....((((((..............))))))...-....))))). ( -27.54, z-score = -2.02, R) >droSec1.super_94 86407 99 - 93855 --------------------ACCACUUGACCACUCCAGCCAUAGUCGCCGAUCGGGGCCACCACCUGGUCCUGAUCUGCACUCUGUACCACACCACAGCUACAGAGACG-UCAACGAAGC --------------------.....(((((................((.(((((((((((.....))))))))))).)).(((((((.(........).)))))))..)-))))...... ( -34.50, z-score = -3.67, R) >droYak2.chrU 2674181 94 - 28119190 -------------------------UUGGUCACCUGAGCUACAGUCACCUAUCGGGGCCACCACCUCGACCCGAUCCGCACUCAGUACCACACCAGAGCCACGGAUACG-ACAACAAGGU -------------------------..(.((....)).).......((((.((((((......))))))..(((((((..(((............)))...))))).))-......)))) ( -20.40, z-score = 0.56, R) >droAna3.scaffold_13417 3988958 120 - 6960332 ACACCUUUAUUAUAACAGCUAAGACCCUGCUAUCAGCACCAUCACCAUUGAUUGUGGCCAUCACCUGGACCCGAUCCGCACUCUGAAUCACUCUACAGUUGCAGAGGCAUUCAACAUAGU .................((((.((...(((.....)))...))......(((((.(((((.....))).))))))).((.(((((....(((....)))..)))))))........)))) ( -23.10, z-score = -0.57, R) >consensus ____________________ACCACUUGGUCACUUCAGCUAUAGUCACCGAUCGGGGCCACCACCUGGUCCUGAUCCGCACUCUGUACCACACCACAGCUACAGAGACG_UCAACGAGGU ..............................................((((((((((((((.....)))))))))))....(((((((.(........).)))))))...........))) (-17.16 = -18.36 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:59:29 2011