| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 728,042 – 728,105 |
| Length | 63 |
| Max. P | 0.924488 |

| Location | 728,042 – 728,105 |
|---|---|
| Length | 63 |
| Sequences | 4 |
| Columns | 64 |
| Reading direction | reverse |
| Mean pairwise identity | 69.55 |
| Shannon entropy | 0.49349 |
| G+C content | 0.36515 |
| Mean single sequence MFE | -11.47 |
| Consensus MFE | -5.00 |
| Energy contribution | -4.88 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.57 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.924488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
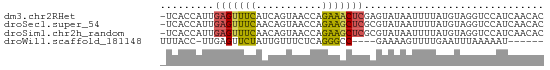
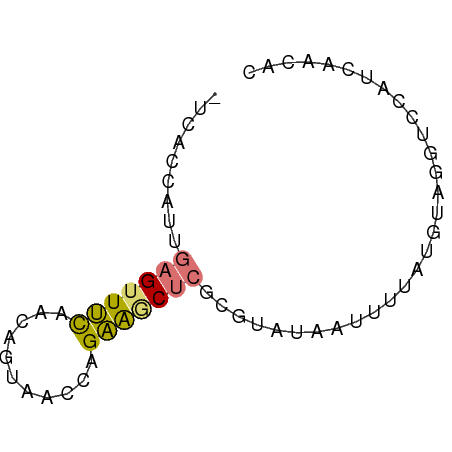
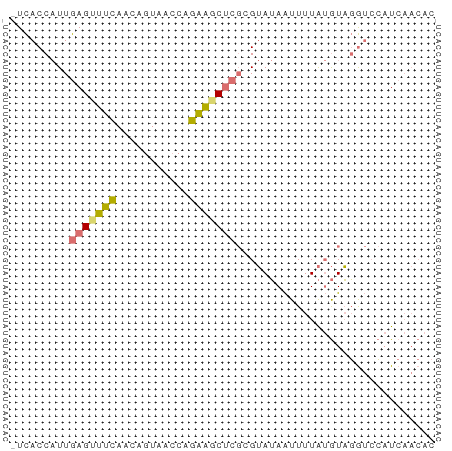
>dm3.chr2RHet 728042 63 - 3288761 -UCACCAUUGAGUUUCAUCAGUAACCAGAAACUCGAGUAUAAUUUUAUGUAGGUCCAUCAACAC -..(((.(((((((((...........))))))))).((((......))))))).......... ( -10.90, z-score = -1.94, R) >droSec1.super_54 165285 63 - 216317 -UCACCAUUGAGUUUCAACAGUAACCAGAAGCUCGCGUAUAAUUUUAUGUAGGUCCAUCAACAC -..(((...(((((((...........)))))))(((((......))))).))).......... ( -13.00, z-score = -3.04, R) >droSim1.chr2h_random 1890332 63 - 3178526 -UCACCAUUGAGUUUCAACAGUAACCAGAAGCUCGCGUAUAAUUUUAUGUAGGUCCAUCAACAC -..(((...(((((((...........)))))))(((((......))))).))).......... ( -13.00, z-score = -3.04, R) >droWil1.scaffold_181148 801862 53 + 5435427 UUUACC-UUGAGUUCUAUUGUUUCUCAGGGCC----GAAAAGUUUUGAAUUUAAAAAU------ ....((-(((((..(....)...)))))))..----......................------ ( -9.00, z-score = -0.86, R) >consensus _UCACCAUUGAGUUUCAACAGUAACCAGAAGCUCGCGUAUAAUUUUAUGUAGGUCCAUCAACAC .........(((((((...........))))))).............................. ( -5.00 = -4.88 + -0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:59:20 2011