
| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 630,539 – 630,630 |
| Length | 91 |
| Max. P | 0.999955 |

| Location | 630,539 – 630,630 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 51.35 |
| Shannon entropy | 0.77710 |
| G+C content | 0.45687 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -14.91 |
| Energy contribution | -15.47 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.62 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 5.19 |
| SVM RNA-class probability | 0.999955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
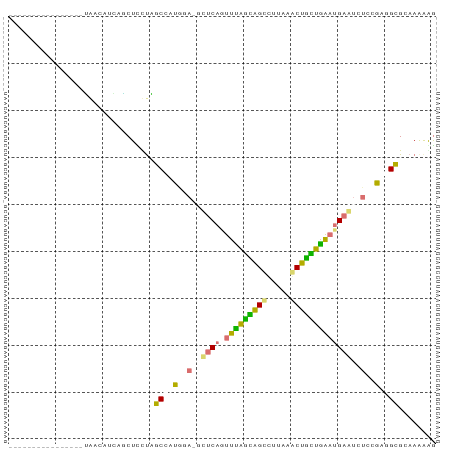
>dm3.chr2RHet 630539 91 + 3288761 UAGUUUUCGUCGAGCCGAUGAUCUUCUUUCCGCGCUUAAUUCACAGUUUAAUGGCCUUAAGCUAUUAGAAGAAUCUCCAAAGCGCAAAAAA ......((((((...))))))..........((((((.((((....(((((((((.....))))))))).)))).....))))))...... ( -22.00, z-score = -1.94, R) >dp4.Unknown_group_58 40948 75 + 69560 ----------------UACCUUCGGCUCCUAGCCAUAGAAGCUCAGUUUAGCGGUUUAAAACUGCUGAAUGAGUCUCCGAGGUGCAAAAAG ----------------((((((.(((.....)))...((..((((.(((((((((.....)))))))))))))..)).))))))....... ( -27.60, z-score = -3.83, R) >droPer1.super_87 105076 72 + 209488 ----------------UGGCUUGCACUCCUAGCCAUGU---CUCAGUUUGGCAGCCUGCAACUGCUGAAUGAGUCUCCGAGGCGUAAAAGG ----------------.((((.........))))((((---(((.(((..((((.......))))..)))(......))))))))...... ( -21.00, z-score = -0.00, R) >droWil1.scaffold_181155 684691 85 - 3442787 ------UCAUAAGCUCCAAGAGGAGCUUUUGGCAAUGGAGGUUCAAUUCAGAAGUUUUAAACUUUUGAUUGAAUCUCCGACGCGAAAGGGG ------(((.(((((((....))))))).)))...((((((((((((.(((((((.....))))))))))))))))))).(.(....).). ( -37.90, z-score = -6.32, R) >consensus ________________UAACAUCAGCUCCUAGCCAUGGA_GCUCAGUUUAGCAGCCUUAAACUGCUGAAUGAAUCUCCGAGGCGCAAAAAG ...............................(((.((((.(((((.(((((((((.....)))))))))))))).)))).)))........ (-14.91 = -15.47 + 0.56)


| Location | 630,539 – 630,630 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 51.35 |
| Shannon entropy | 0.77710 |
| G+C content | 0.45687 |
| Mean single sequence MFE | -21.88 |
| Consensus MFE | -9.94 |
| Energy contribution | -11.75 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.986386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
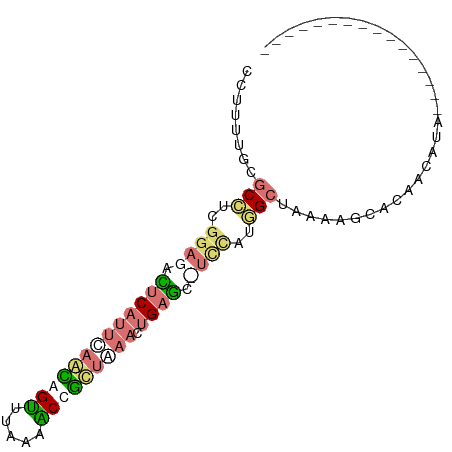
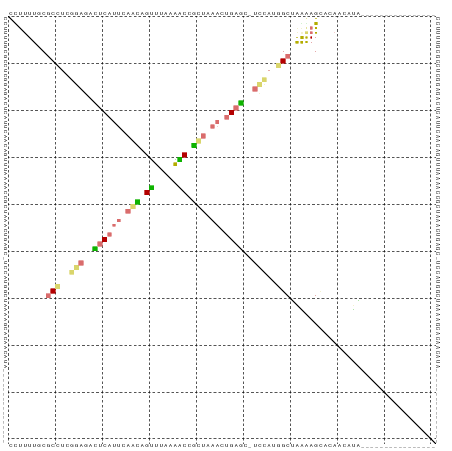
>dm3.chr2RHet 630539 91 - 3288761 UUUUUUGCGCUUUGGAGAUUCUUCUAAUAGCUUAAGGCCAUUAAACUGUGAAUUAAGCGCGGAAAGAAGAUCAUCGGCUCGACGAAAACUA ((((((.(((.((((((....))))))..((((((..(((......)).)..))))))))).)))))).....(((......)))...... ( -20.70, z-score = -0.40, R) >dp4.Unknown_group_58 40948 75 - 69560 CUUUUUGCACCUCGGAGACUCAUUCAGCAGUUUUAAACCGCUAAACUGAGCUUCUAUGGCUAGGAGCCGAAGGUA---------------- ........(((((((((.((((((.(((.((.....)).))).)).))))))))...(((.....)))).)))).---------------- ( -19.70, z-score = -0.72, R) >droPer1.super_87 105076 72 - 209488 CCUUUUACGCCUCGGAGACUCAUUCAGCAGUUGCAGGCUGCCAAACUGAG---ACAUGGCUAGGAGUGCAAGCCA---------------- .((((((.(((..(....((((((..(((((.....)))))..)).))))---.)..))))))))).........---------------- ( -17.10, z-score = 1.22, R) >droWil1.scaffold_181155 684691 85 + 3442787 CCCCUUUCGCGUCGGAGAUUCAAUCAAAAGUUUAAAACUUCUGAAUUGAACCUCCAUUGCCAAAAGCUCCUCUUGGAGCUUAUGA------ ........(((..((((.((((((((.((((.....)))).)).)))))).))))..)))((.(((((((....))))))).)).------ ( -30.00, z-score = -5.86, R) >consensus CCUUUUGCGCCUCGGAGACUCAUUCAACAGUUUAAAACCGCUAAACUGAGC_UCCAUGGCUAAAAGCACAACAUA________________ ........(((..((((.(((((((((((((.....))))))))).)))).))))..)))............................... ( -9.94 = -11.75 + 1.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:59:19 2011