
| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 574,896 – 574,946 |
| Length | 50 |
| Max. P | 0.500000 |

| Location | 574,896 – 574,946 |
|---|---|
| Length | 50 |
| Sequences | 5 |
| Columns | 59 |
| Reading direction | forward |
| Mean pairwise identity | 52.05 |
| Shannon entropy | 0.78482 |
| G+C content | 0.37076 |
| Mean single sequence MFE | -7.18 |
| Consensus MFE | -2.14 |
| Energy contribution | -1.86 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 2.00 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
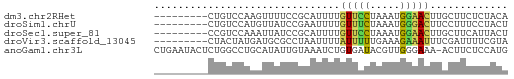
>dm3.chr2RHet 574896 50 + 3288761 ---------CUGUCCAAGUUUUCCGCAUUUUGUUCCUAAAUGGAACUUGCUUCUCUACA ---------...............(((....(((((.....))))).)))......... ( -9.60, z-score = -2.67, R) >droSim1.chrU 5054032 50 + 15797150 ---------CUGUCCAUGUUAUCCGAAUUUUGUUUCUAAAUGGGACUUCCUUUCCUACU ---------..((((...(((...(((......))))))...))))............. ( -6.10, z-score = -0.56, R) >droSec1.super_81 28126 50 + 123496 ---------CCGUCCAAAUUAUCCGCAUUUUGUUCCUAAAUGGAACUUGCUUCAUUACU ---------...............(((....(((((.....))))).)))......... ( -9.60, z-score = -3.41, R) >droVir3.scaffold_13045 1529520 50 - 2268007 ---------CUACUAUGAUGCGCCUAAUUUUAUUUUUGAAAGAAAUUUCGAUUUUCGUA ---------..........................((((((....))))))........ ( -4.70, z-score = -0.11, R) >anoGam1.chr3L 35439481 58 - 41284009 CUGAAUACUCUGGCCUGCAUAUUGUAAAUCUGUGAUACGUUGGGAAA-ACUUCUCCAUG .............((.((.(((..((....))..))).)).))....-........... ( -5.90, z-score = 0.74, R) >consensus _________CUGUCCAAAUUAUCCGAAUUUUGUUCCUAAAUGGAACUUCCUUCUCUACA ...............................(((((.....)))))............. ( -2.14 = -1.86 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:59:15 2011