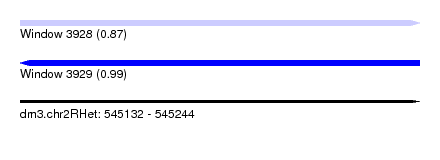
| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 545,132 – 545,244 |
| Length | 112 |
| Max. P | 0.993970 |

| Location | 545,132 – 545,244 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 128 |
| Reading direction | forward |
| Mean pairwise identity | 55.05 |
| Shannon entropy | 0.71299 |
| G+C content | 0.46430 |
| Mean single sequence MFE | -33.44 |
| Consensus MFE | -17.71 |
| Energy contribution | -18.15 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.20 |
| Mean z-score | -0.66 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.872217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
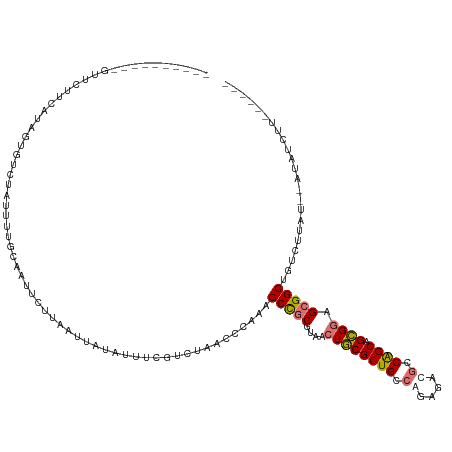
>dm3.chr2RHet 545132 112 + 3288761 ----------CUUCUCCACAGUGUCUGUGUUGUGAGUCUUAAAUAAAUUUCGUCUUAGGCUAAGCCGCGUAACUGCGCUGCCGGAUACGCCAGCGGCGUAGCGGCUUCCUUAUGUAUAUCUU------ ----------...((((((((...)))))....)))..............(((...(((..(((((((.....((((((((.((.....)).)))))))))))))))))).)))........------ ( -32.40, z-score = -0.95, R) >droYak2.chrX_random 539130 108 + 1802292 ----------UUUCUACA-AGUGUCUGUUUUGUGAUUCUUAAUUAUAUUUCGUCGAAGGCUAAGCCGCGUAGCUGCGCUGCCGGAUACGACAGCAGCGGAGCGGCUCUCUUA---AUAUCUU------ ----------((((.(((-(((((..(((..(.....)..))).)))))).)).))))....((((((....((((((((.((....)).)))).)))).))))))......---.......------ ( -29.30, z-score = -0.04, R) >droEre2.scaffold_4929 25605958 117 + 26641161 ---UUUCUGUGGUCUUUAUUAUAUUCAUCUUCCAAAAAGUACUUAUCUUUAAAUUAGCCUAGAGCUGCGCAACUUCGCUGCCAGAGAGACCGGCAGGGGAGGGGCUGUCUUAU--CUAUUUU------ ---.(((((.(((.((((..........(((.....)))..........))))...))))))))....((..((((.(((((.........))))).))))..))........--.......------ ( -23.55, z-score = 1.56, R) >droPer1.super_1295 3240 128 - 6930 CUGUUUUAUGGUGAUUUCUCUUCUAUAUUUUACAAUUGUGGUUUGGGCUUUUGUAAGCCCAAAGCCGCGGAGCUGCGCUGCCAGUGACGCCGGCAGCGGAGCGGCAGAACUAUGUGUUCUUAUCUCUU .(((..(((((.((....))..)))))....))).(((((((((((((((....)))))).))))))))).(((.(((((((.(.....).))))))).)))...(((((.....)))))........ ( -48.50, z-score = -3.23, R) >consensus __________GUUCUUCAUAGUGUCUAUUUUGCAAUUCUUAAUUAUAUUUCGUCUAACCCAAAGCCGCGUAACUGCGCUGCCAGAGACGCCAGCAGCGGAGCGGCUGUCUUAU__AUAUCUU______ ...............................................................(((((....((((((((.((....)).)))).)))).)))))....................... (-17.71 = -18.15 + 0.44)

| Location | 545,132 – 545,244 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 128 |
| Reading direction | reverse |
| Mean pairwise identity | 55.05 |
| Shannon entropy | 0.71299 |
| G+C content | 0.46430 |
| Mean single sequence MFE | -30.43 |
| Consensus MFE | -18.09 |
| Energy contribution | -19.52 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.993970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
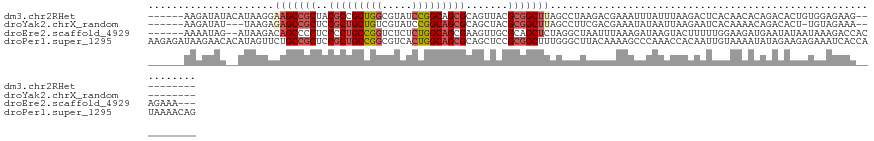
>dm3.chr2RHet 545132 112 - 3288761 ------AAGAUAUACAUAAGGAAGCCGCUACGCCGCUGGCGUAUCCGGCAGCGCAGUUACGCGGCUUAGCCUAAGACGAAAUUUAUUUAAGACUCACAACACAGACACUGUGGAGAAG---------- ------............(((.((((((.((..(((((.((....)).)))))..))...))))))...)))....................(((....(((((...))))))))...---------- ( -31.80, z-score = -1.85, R) >droYak2.chrX_random 539130 108 - 1802292 ------AAGAUAU---UAAGAGAGCCGCUCCGCUGCUGUCGUAUCCGGCAGCGCAGCUACGCGGCUUAGCCUUCGACGAAAUAUAAUUAAGAAUCACAAAACAGACACU-UGUAGAAA---------- ------.......---.....(((((((...((((((((((....)))))..)))))...)))))))....................((((..((........))..))-))......---------- ( -26.70, z-score = -0.50, R) >droEre2.scaffold_4929 25605958 117 - 26641161 ------AAAAUAG--AUAAGACAGCCCCUCCCCUGCCGGUCUCUCUGGCAGCGAAGUUGCGCAGCUCUAGGCUAAUUUAAAGAUAAGUACUUUUUGGAAGAUGAAUAUAAUAAAGACCACAGAAA--- ------.......--.......((((.......((((((.....))))))(((......))).......))))..((((((((.......))))))))...........................--- ( -19.40, z-score = 1.51, R) >droPer1.super_1295 3240 128 + 6930 AAGAGAUAAGAACACAUAGUUCUGCCGCUCCGCUGCCGGCGUCACUGGCAGCGCAGCUCCGCGGCUUUGGGCUUACAAAAGCCCAAACCACAAUUGUAAAAUAUAGAAGAGAAAUCACCAUAAAACAG ........(((((.....)))))((.(((.(((((((((.....))))))))).)))...))((.(((((((((....)))))))))))...................((....))............ ( -43.80, z-score = -4.46, R) >consensus ______AAAAUAU__AUAAGACAGCCGCUCCGCUGCCGGCGUAUCCGGCAGCGCAGCUACGCGGCUUAGCCCUAAACGAAAUAUAAUUAAGAAUCACAAAACAGACACUAUAAAGAAC__________ .....................(((((((..(((((((((.....))))))))).......)))))))............................................................. (-18.09 = -19.52 + 1.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:59:12 2011