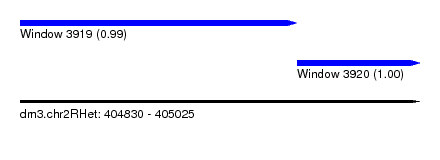
| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 404,830 – 405,025 |
| Length | 195 |
| Max. P | 0.996957 |

| Location | 404,830 – 404,965 |
|---|---|
| Length | 135 |
| Sequences | 3 |
| Columns | 149 |
| Reading direction | forward |
| Mean pairwise identity | 61.89 |
| Shannon entropy | 0.50985 |
| G+C content | 0.42540 |
| Mean single sequence MFE | -41.63 |
| Consensus MFE | -16.58 |
| Energy contribution | -15.60 |
| Covariance contribution | -0.98 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.990568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
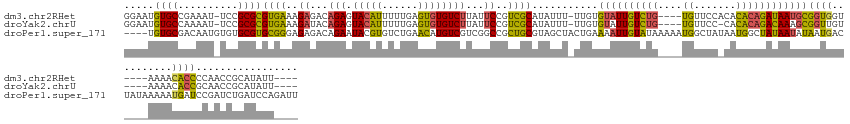
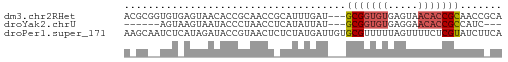
>dm3.chr2RHet 404830 135 + 3288761 GGAAUGUGCCGAAAU-UCCGCGCGUGAAAGAGACAGAGUACAUUUUUGAGUGUGUCUUAUUCCGUCGCAUAUUU-UUGUGUAUUGUCUG----UGUUCCACACACAGAUAAUGCGGUGGU----AAAACACCCCAACCGCAUAUU---- ..((((((((((((.-...(((((.(((.((((((.(.(.((....))).).)))))).))))).)))....))-))).((((((((((----(((.....)))))))))))))((((..----....))))......)))))))---- ( -47.90, z-score = -4.74, R) >droYak2.chrU 13921229 134 - 28119190 GGAAUGUGCCAAAAU-UCCGCGCGUGAAAGAUACAGAGUACAUUUUUGAGUGUGUCUUAUUCCGUCGCAUAUUU-UUGUGUAUUGUCUG----UGUUCC-CACACAGACAAAGCGGUUGU----AAAACACCGCAACCGCAUAUU---- (((((........))-)))(((((.(((((((((((((......)))...)))))))..))))).)))......-.......(((((((----(((...-.)))))))))).((((((((----........)))))))).....---- ( -45.40, z-score = -4.49, R) >droPer1.super_171 24541 145 + 68591 ----UGUGCGACAAUGUGUGCGUGCGGGAGAGACAGAAUACGUGUCUGAACAUGUCGUCGGCCGCUGCGUAGCUACUGAAAAUUGUAUAAAAAUGGCUAUAAUGGCUAUAAUAUAAUGACUAUAAAAAUGAUCCGAUCUGAUCCAGAUU ----.(..((((.....)).))..).(((.(((((.......)))))......(((.((((..(((....)))..))))..(((((((....((((((.....)))))).))))))))))...........)))((((((...)))))) ( -31.60, z-score = 0.34, R) >consensus GGAAUGUGCCAAAAU_UCCGCGCGUGAAAGAGACAGAGUACAUUUUUGAGUGUGUCUUAUUCCGUCGCAUAUUU_UUGUGUAUUGUCUG____UGUUCC_CACACAGAUAAUGCGGUGGU____AAAACACCCCAACCGCAUAUU____ .....((((..........))))((((..((...(((.(((((......))))))))...))..))))...........((((((((((....((.......))))))))))))................................... (-16.58 = -15.60 + -0.98)
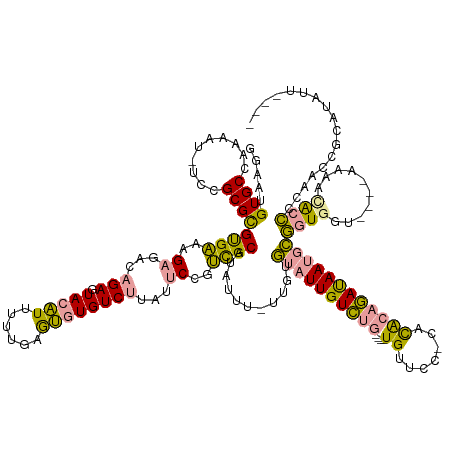
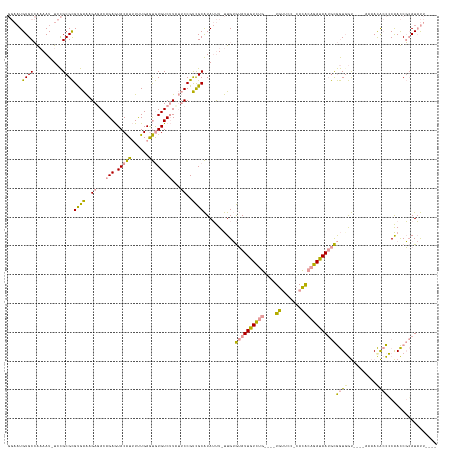
| Location | 404,965 – 405,025 |
|---|---|
| Length | 60 |
| Sequences | 3 |
| Columns | 63 |
| Reading direction | forward |
| Mean pairwise identity | 52.69 |
| Shannon entropy | 0.66111 |
| G+C content | 0.45562 |
| Mean single sequence MFE | -16.77 |
| Consensus MFE | -7.22 |
| Energy contribution | -8.33 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.01 |
| SVM RNA-class probability | 0.996957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
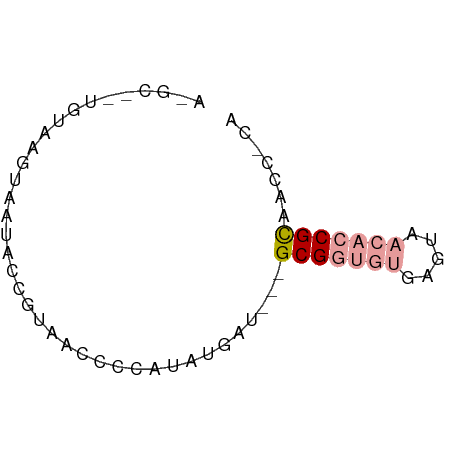
>dm3.chr2RHet 404965 60 + 3288761 ACGCGGUGUGAGUAACACCGCAACCGCAUUUGAU---GCGGUGUGAGUAACACCGCAACCGCA ..(((((((.....)))))))....((....(.(---(((((((.....)))))))).).)). ( -27.90, z-score = -3.44, R) >droYak2.chrU 9225789 51 + 28119190 ------AGUAAGUAAUACCCUAACCUCAUAUUAU---GCGGUGUGAGGAACACCGCCAUC--- ------............................---(((((((.....)))))))....--- ( -12.20, z-score = -1.83, R) >droPer1.super_171 24686 63 + 68591 AAGCAAUCUCAUAGAUACCGUAACUCUCUAUGAUUGUGCGUUUUUAGUUUUCUCGUAUCUUCA ..(((...(((((((...........)))))))...)))........................ ( -10.20, z-score = -1.98, R) >consensus A_GC__UGUAAGUAAUACCGUAACCCCAUAUGAU___GCGGUGUGAGUAACACCGCAACC_CA .....................................(((((((.....)))))))....... ( -7.22 = -8.33 + 1.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:59:04 2011