
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,060,252 – 2,060,368 |
| Length | 116 |
| Max. P | 0.714436 |

| Location | 2,060,252 – 2,060,368 |
|---|---|
| Length | 116 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.66 |
| Shannon entropy | 0.39906 |
| G+C content | 0.42263 |
| Mean single sequence MFE | -28.64 |
| Consensus MFE | -17.50 |
| Energy contribution | -16.80 |
| Covariance contribution | -0.70 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.714436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
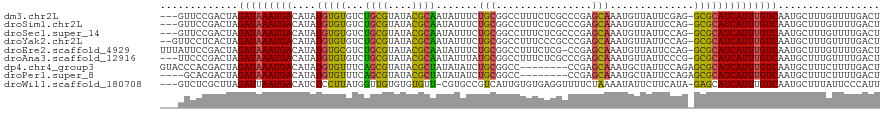
>dm3.chr2L 2060252 116 + 23011544 ---GUUCCGACUAGAUAAAUGACAUAUGUGUGUCUGCGUAUACGCAAUAUUUCUGCGGCCUUUCUCGCCCGAGCAAAUGUUAUUCGAG-GCGCAUCAUUUGUCAAUGCUUUGUUUUGACU ---(((..(((.((...(.(((((.(((((((((((((....)))(((((((((.(((.(......).))))).))))))).....))-))))).))).))))).).))..)))..))). ( -31.00, z-score = -1.53, R) >droSim1.chr2L 2030438 116 + 22036055 ---GUUCCGACUAGAUAAAUGACAUAUGUGUGUCUGCGUAUACGCAAUAUUUCUGCGGCCUUUCUCGCCCGAGCAAAUGUUAUUCCAG-GCGCAUCAUUUGUCAAUGCUUUGUUUUGACU ---(((..(((.((...(.(((((.(((((((((((((....)))(((((((((.(((.(......).))))).))))))).....))-))))).))).))))).).))..)))..))). ( -30.90, z-score = -1.87, R) >droSec1.super_14 2006808 116 + 2068291 ---GUUCCGACUAGAUAAAUGACAUAUGUGUGUCUGCGUAUACGCAAUAUUUCUGCGGCCUUUCUCGCCCGAGCAAAUGUUAUUCCAG-GCGCAUCAUUUGUCAAUGCUUUGUUUUGACU ---(((..(((.((...(.(((((.(((((((((((((....)))(((((((((.(((.(......).))))).))))))).....))-))))).))).))))).).))..)))..))). ( -30.90, z-score = -1.87, R) >droYak2.chr2L 2046585 117 + 22324452 --GUUCCUCACUAGAUAAAUGACAUAUGUGUGUCUGCGUAUACGCAAUAUUUCUGCGGCCUUUCCCGCCCGAGCAAAUGUUAUUCCAG-GCGCAUCAUUUGUCAAUGCUUUGUUUUGACU --.....(((..((((((((((((.(((((((((((((....)))(((((((((((((......))))...)).))))))).....))-))))).))).)))))....)))))))))).. ( -31.10, z-score = -2.36, R) >droEre2.scaffold_4929 2108571 118 + 26641161 UUUAUUCCGACUAGAUAAAUGACAUAUGUGCGUCUGCGUAUACGCAAUAUUUCUGCGGCCUUUCUCG-CCGAGCAAAUGUUAUUCCAG-GCGCAUCAUUUGUCAAUGCUUUGUUUUGACU ..........(.((((((((((((.(((((((((((((....)))(((((((((.((((.......)-))))).))))))).....))-))))).))).)))))....))))))).)... ( -34.70, z-score = -2.99, R) >droAna3.scaffold_12916 9775176 116 - 16180835 ---UUCCCGACUAGAUAAAUGACAUAUGUGUGUCUGCGUAUACGCAAUAUUUAUGCGGCCUUUCUCGCCCGAGCAAAUGUUAUUCCCG-GCGCAUCAUUUGUCAAUGCUUUGUUUUGACU ---.......(.((((((((((((.(((((((((.(((....)))(((((((...(((.(......).)))...)))))))......)-))))).))).)))))....))))))).)... ( -28.00, z-score = -1.28, R) >dp4.chr4_group3 8859338 112 + 11692001 GUACCCACGACUAGAUAAAUGACAUAUGUGUUUCAGCGUAUACGCUAUAUAUCUGCGGCC--------CCGAGCAAAUGCUAUUCCAGAGCGCAUCAUUUGUCAAUGCUUUCUUUUGACU .............(((((((((....(((((...((((....)))).....((((((...--------.))(((....)))....))))))))))))))))))................. ( -24.60, z-score = -0.84, R) >droPer1.super_8 41552 108 + 3966273 ----GCACGACUAGAUAAAUGACAUAUGUGUUUCAGCGUAUACGCUAUAUAUCUGCGGCC--------CCGAGCAAAUGCUAUUCCAGAGCGCAUCAUUUGUCAAUGCUUUCUUUUGACU ----.........(((((((((....(((((...((((....)))).....((((((...--------.))(((....)))....))))))))))))))))))................. ( -24.60, z-score = -0.54, R) >droWil1.scaffold_180708 11722521 115 - 12563649 ---GUCUCGCUUAGAUUAAUGACAUCUCCUUAUGGUUGUGUGUGUU-CGUGCCGUCAUUGUGUGAGGUUUUCUAAAAUAUUCUUCAUA-GAGCAUCAUUUGUCAAUGCUUUAUUCCCAUU ---.((((((......(((((((....((....))..(..((....-))..).))))))).))))))..................(((-((((((.........)))))))))....... ( -22.00, z-score = -0.94, R) >consensus ___GUCCCGACUAGAUAAAUGACAUAUGUGUGUCUGCGUAUACGCAAUAUUUCUGCGGCCUUUCUCGCCCGAGCAAAUGUUAUUCCAG_GCGCAUCAUUUGUCAAUGCUUUGUUUUGACU .............(((((((((....(((((...((((....)))).........(((..........)))..................))))))))))))))................. (-17.50 = -16.80 + -0.70)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:10:05 2011