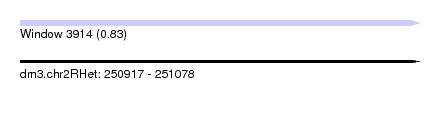
| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 250,917 – 251,078 |
| Length | 161 |
| Max. P | 0.826615 |

| Location | 250,917 – 251,078 |
|---|---|
| Length | 161 |
| Sequences | 4 |
| Columns | 171 |
| Reading direction | forward |
| Mean pairwise identity | 66.43 |
| Shannon entropy | 0.53409 |
| G+C content | 0.44867 |
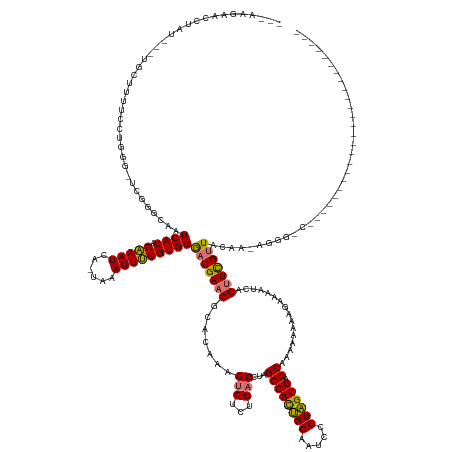
| Mean single sequence MFE | -41.45 |
| Consensus MFE | -20.79 |
| Energy contribution | -20.23 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.826615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
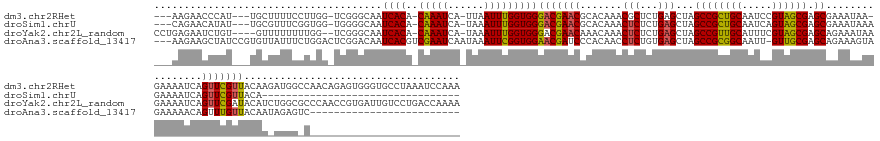
>dm3.chr2RHet 250917 161 + 3288761 ---AAGAACCCAU---UGCUUUUCCUUGG-UCGGGCAAUCACA-CAAAUCA-UUAAUUUGGUGGGACGAACGCACAAACGCUCUGAGCUAGCCGCUGCAAUCCGUAGCGAGCGAAAUAA-GAAAAUCAGUUCGUUACAAGAUGGCCAACAGAGUGGGUGCCUAAAUCCAAA ---........((---(((((..(....)-..)))))))(((.-(((((..-...))))))))(((.....((((...((((((((((.....)))((.(((.(((((((((.......-........)))))))))..))).))...))))))).)))).....)))... ( -47.96, z-score = -1.72, R) >droSim1.chrU 2373484 129 - 15797150 ---CAGAACAUAU---UGCGUUUCGGUGG-UGGGGCAAUCACA-CAAAUCA-UAAAUUUGGUGGGACGAACGCACAAACUCUCUGAGCUAGCCGCUGCAAUCAGUAGCGAGCGAAAUAAAGAAAAUCAGUUCGUUACA--------------------------------- ---..((((....---((((((.((((((-(......))))).-(..((((-......))))..).)))))))).......(((......((((((((.....)))))).)).......)))......))))......--------------------------------- ( -34.62, z-score = -1.48, R) >droYak2.chr2L_random 977845 163 - 4064425 CCUGAGAAUCUGU----GUUUUUUUUGG--UCGGGCAAUCACA-CAAAUCA-UAAAUUUGGUGGGACGAACAAACAAACUCUCUGAGCUAGCCGUUGCAUUUCGUAGCGAGCAGAAAUAAGAAAAUCAGUUCGAUACAUCUGGCGCCCAACCGUGAUUGUCCUGACCAAAA .............----.....((((((--((((((((((((.-(((((..-...))))).((((.((.............(((...((.((((((((.....)))))).)))).....)))...((((..........))))))))))...))))))).))))))))))) ( -48.60, z-score = -2.46, R) >droAna3.scaffold_13417 3433053 142 + 6960332 ---AAGAAGCUAUCCGUGUUAUUUCUGGACUCGGACAAUCACGUCGAAUCAAUAAAUUCGGUGGAACGAUCCCACAACCUCUGUGAGCUAGCCGCGGCAAUU-GUUGCGAGCAGAAAGUAGAAAAACAGUUUGUUACAAUAGAGUC------------------------- ---.....((((((((.(((.......))).))))...((((..(((((......)))))((((.......)))).......))))..))))((((((....-))))))((((((..((......))..))))))...........------------------------- ( -34.60, z-score = -0.31, R) >consensus ___AAGAACCUAU___UGCUUUUCCUGGG_UCGGGCAAUCACA_CAAAUCA_UAAAUUUGGUGGGACGAACGCACAAACUCUCUGAGCUAGCCGCUGCAAUCCGUAGCGAGCAAAAAAAAGAAAAUCAGUUCGUUACAA_AGGG_C_________________________ ......................................((((..(((((......)))))))))(((((((.......(((...)))...((((((((.....)))))).))................))))))).................................... (-20.79 = -20.23 + -0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:59:00 2011