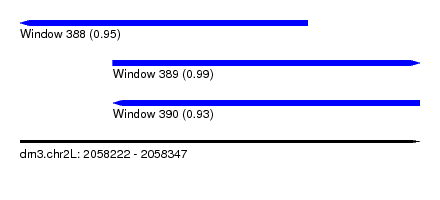
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,058,222 – 2,058,347 |
| Length | 125 |
| Max. P | 0.992592 |

| Location | 2,058,222 – 2,058,312 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 69.02 |
| Shannon entropy | 0.51643 |
| G+C content | 0.56957 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -14.41 |
| Energy contribution | -13.64 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.954234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
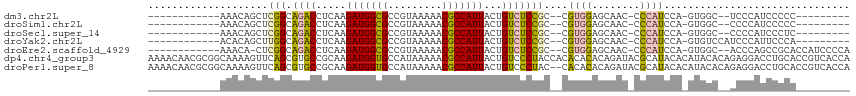
>dm3.chr2L 2058222 90 - 23011544 ------------AAACAGCUCGGCAGACCUCAAGAUGGCGCCGUAAAAACGCCAUUACUGUCUCCGC--CGUGGAGCAAC-CCCAUCCA-GUGGC--UCCCAUCCCCC--------- ------------........((((((((.....(((((((.........)))))))...))))..))--)).(((((.((-........-)).))--)))........--------- ( -26.00, z-score = -2.13, R) >droSim1.chr2L 2028471 90 - 22036055 ------------AAACAGCUCGGCAGACCUCAAGAUGGCGCCGUAAAAACGCCAUUACUGUCUCCGC--CGUGGAGCAAC-CCCAUCCA-GUGGC--CCCCAUCCCCC--------- ------------.........((.((((.....(((((((.........)))))))...))))))((--((((((.....-....))))-.))))--...........--------- ( -24.20, z-score = -1.55, R) >droSec1.super_14 2005010 90 - 2068291 ------------AAACAGCUCGGCAGACCUCAAGAUGGCGCCGUAAAAACGCCAUUACUGUCUCCGC--CGUGGAGCAAC-CCCAUCCA-GUGGC--CCCCAUCCCUC--------- ------------.........((.((((.....(((((((.........)))))))...))))))((--((((((.....-....))))-.))))--...........--------- ( -24.20, z-score = -1.47, R) >droYak2.chr2L 2044302 92 - 22324452 ------------ACACAGCUUGGCAGACCUCAAGAUGGCGCCGUAAAAACGCCAUUACUGUCUCCGC--CGUGGAGCAAC-CCCAUCCA-GUGUCCAUCCCAUUCCCA--------- ------------((((.....(((((((.....(((((((.........)))))))...))))..))--).((((.....-....))))-))))..............--------- ( -22.50, z-score = -1.36, R) >droEre2.scaffold_4929 2106382 98 - 26641161 ------------AAACA-CUCGGCAGACCUCAAGAUGGCGCCGUAAAAACGCCAUUACUGUCUCCGC--CGUGGAGCAAC-CCCAUCCA-GUGGC--ACCCAGCCGCACCAUCCCCA ------------.....-..((((((((.....(((((((.........)))))))...))))..))--))((((.....-....))))-(((((--.....))))).......... ( -28.90, z-score = -2.43, R) >dp4.chr4_group3 8856264 117 - 11692001 AAAACAACGCGGCAAAAGUUCAGCGUGCCGCAAGAUGGUGCCAUAAAAACGCCAUUACUGUCCCUACCACACACACAGAUACGCAUACACAUACACAGAGGACCUGCACCGUCACCA ........((((((...(.....).))))))..((((((((.........((.....((((.............))))....))............((.....)))))))))).... ( -27.02, z-score = -1.56, R) >droPer1.super_8 38474 115 - 3966273 AAAACAACGCGGCAAAAGUUCAGCGUGCCGCAAGAUGGUGCCAUAAAAACGCCAUUACUGUCCCUAC--CACACACAGAUACGCAUACACAUACACAGAGGACCUGCACCGUCACCA ........((((((...(.....).))))))..((((((((.........((.....((((......--.....))))....))............((.....)))))))))).... ( -27.20, z-score = -1.66, R) >consensus ____________AAACAGCUCGGCAGACCUCAAGAUGGCGCCGUAAAAACGCCAUUACUGUCUCCGC__CGUGGAGCAAC_CCCAUCCA_GUGGC__CCCCAUCCCCA_________ ....................(((.((((.....(((((((.........)))))))...)))))))....((((........))))............................... (-14.41 = -13.64 + -0.77)
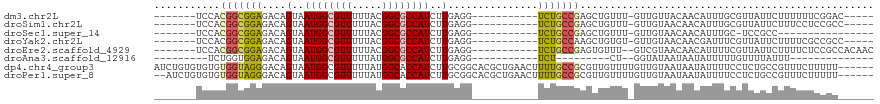
| Location | 2,058,251 – 2,058,347 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.74 |
| Shannon entropy | 0.55554 |
| G+C content | 0.46801 |
| Mean single sequence MFE | -31.01 |
| Consensus MFE | -14.34 |
| Energy contribution | -14.38 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.992592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
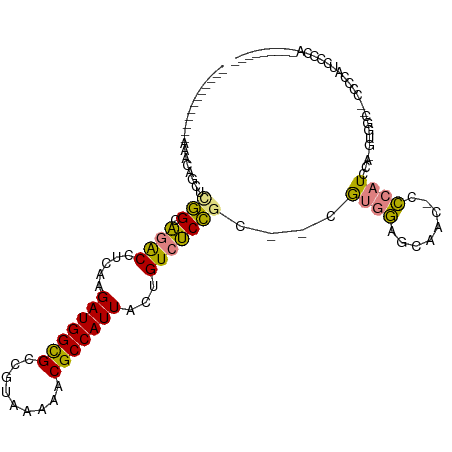
>dm3.chr2L 2058251 96 + 23011544 -------UCCACGGCGGAGACAGUAAUGGCGUUUUUACGGCGCCAUCUUGAGG-----------UCUGCCGAGCUGUUU-GUUGUUACAACAUUUGCGUUAUUCUUUUUUCGGAC----- -------(((.(((((((....(..((((((((.....))))))))..)....-----------))))))).((....(-((((...)))))...))..............))).----- ( -30.60, z-score = -2.13, R) >droSim1.chr2L 2028500 96 + 22036055 -------UCCACGGCGGAGACAGUAAUGGCGUUUUUACGGCGCCAUCUUGAGG-----------UCUGCCGAGCUGUUU-GUUGUAACAACAUUUGCGUUAUUCUUUCCUCCGCC----- -------.....((((((((((((.((((((((.....)))))))).....((-----------....))..))))).(-((((...)))))................)))))))----- ( -32.20, z-score = -2.28, R) >droSec1.super_14 2005039 84 + 2068291 -------UCCACGGCGGAGACAGUAAUGGCGUUUUUACGGCGCCAUCUUGAGG-----------UCUGCCGAGCUGUUU-GUUGUAACAACAUUUGC-UCCGCC---------------- -------.....((((((((((((.((((((((.....)))))))).....((-----------....))..))))).(-((((...)))))....)-))))))---------------- ( -32.10, z-score = -2.71, R) >droYak2.chr2L 2044333 96 + 22324452 -------UCCACGGCGGAGACAGUAAUGGCGUUUUUACGGCGCCAUCUUGAGG-----------UCUGCCAAGCUGUGU-GUUGUAACAACGAUUUCGUUAUUCUUUUCGCCGCC----- -------....(((((((((.((((((((((((.(((((((((((.((((...-----------.....)))).)).))-))))))).))))...)))))))).)))))))))..----- ( -38.20, z-score = -3.86, R) >droEre2.scaffold_4929 2106420 100 + 26641161 -------UCCACGGCGGAGACAGUAAUGGCGUUUUUACGGCGCCAUCUUGAGG-----------UCUGCCGAGUGUUU--GUCGUAACAACAUUUUCGUUAUUCUUUUCUCCGCCACAAC -------.....((((((((.((.(((((((......(((((((.......))-----------)..))))((((((.--((....))))))))..))))))).)).))))))))..... ( -34.10, z-score = -3.32, R) >droAna3.scaffold_12916 9773194 75 - 16180835 ---------UCUGGUGGAGACAGUAAUGGCGUUUUUAUGGCGCCAUCUUGAGG-----------UCU---------CU--GGUAUAAUAAUAUUUUUGUUUUAUUU-------------- ---------......((((((.(..((((((((.....))))))))..)...)-----------)))---------))--((((.(((((.....))))).)))).-------------- ( -20.90, z-score = -3.00, R) >dp4.chr4_group3 8856301 114 + 11692001 AUCUGUGUGUGUGGUAGGGACAGUAAUGGCGUUUUUAUGGCACCAUCUUGCGGCACGCUGAACUUUUGCCGCGUUGUUUUGUUGUAAUAAUAUUUUCCUCUGCCGUUUCUUUUU------ ............((((((((((....))..(((.(((..(((..((..(((((((...........)))))))..))..)))..))).)))....))).)))))..........------ ( -30.00, z-score = -1.22, R) >droPer1.super_8 38511 112 + 3966273 --AUCUGUGUGUGGUAGGGACAGUAAUGGCGUUUUUAUGGCACCAUCUUGCGGCACGCUGAACUUUUGCCGCGUUGUUUUGUUGUAAUAAUAUUUUCCUCUGCCGUUUCUUUUU------ --..........((((((((((....))..(((.(((..(((..((..(((((((...........)))))))..))..)))..))).)))....))).)))))..........------ ( -30.00, z-score = -1.45, R) >consensus _______UCCACGGCGGAGACAGUAAUGGCGUUUUUACGGCGCCAUCUUGAGG___________UCUGCCGAGCUGUUU_GUUGUAACAACAUUUUCGUUAUUCUUUUCUCCGC______ ...........(((((((.......((((((((.....))))))))(....)............)))))))................................................. (-14.34 = -14.38 + 0.03)

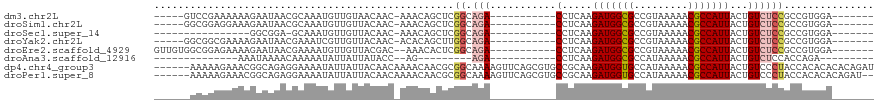
| Location | 2,058,251 – 2,058,347 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.74 |
| Shannon entropy | 0.55554 |
| G+C content | 0.46801 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -8.96 |
| Energy contribution | -9.40 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.928753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
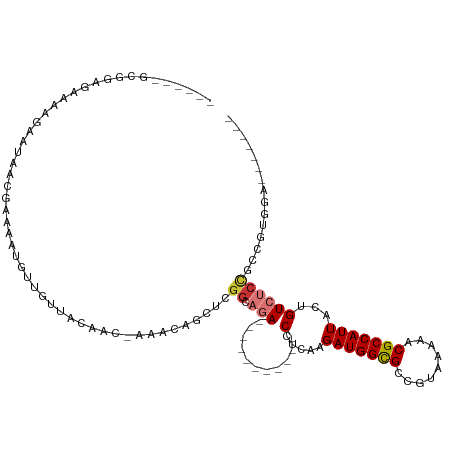
>dm3.chr2L 2058251 96 - 23011544 -----GUCCGAAAAAAGAAUAACGCAAAUGUUGUAACAAC-AAACAGCUCGGCAGA-----------CCUCAAGAUGGCGCCGUAAAAACGCCAUUACUGUCUCCGCCGUGGA------- -----.((((...................(((((......-..))))).(((((((-----------(.....(((((((.........)))))))...))))..))))))))------- ( -23.70, z-score = -1.30, R) >droSim1.chr2L 2028500 96 - 22036055 -----GGCGGAGGAAAGAAUAACGCAAAUGUUGUUACAAC-AAACAGCUCGGCAGA-----------CCUCAAGAUGGCGCCGUAAAAACGCCAUUACUGUCUCCGCCGUGGA------- -----((((((((..((......((....((((((.....-.))))))...))...-----------......(((((((.........))))))).)).)))))))).....------- ( -32.70, z-score = -2.65, R) >droSec1.super_14 2005039 84 - 2068291 ----------------GGCGGA-GCAAAUGUUGUUACAAC-AAACAGCUCGGCAGA-----------CCUCAAGAUGGCGCCGUAAAAACGCCAUUACUGUCUCCGCCGUGGA------- ----------------((((((-((....((((((.....-.))))))...)).((-----------(.....(((((((.........)))))))...))))))))).....------- ( -30.40, z-score = -2.57, R) >droYak2.chr2L 2044333 96 - 22324452 -----GGCGGCGAAAAGAAUAACGAAAUCGUUGUUACAAC-ACACAGCUUGGCAGA-----------CCUCAAGAUGGCGCCGUAAAAACGCCAUUACUGUCUCCGCCGUGGA------- -----.(((((......(((((((....))))))).....-.........((.(((-----------(.....(((((((.........)))))))...)))))))))))...------- ( -30.10, z-score = -1.89, R) >droEre2.scaffold_4929 2106420 100 - 26641161 GUUGUGGCGGAGAAAAGAAUAACGAAAAUGUUGUUACGAC--AAACACUCGGCAGA-----------CCUCAAGAUGGCGCCGUAAAAACGCCAUUACUGUCUCCGCCGUGGA------- .(..(((((((((..((.....(((.....((((....))--))....))).....-----------......(((((((.........))))))).)).)))))))))..).------- ( -31.90, z-score = -2.30, R) >droAna3.scaffold_12916 9773194 75 + 16180835 --------------AAAUAAAACAAAAAUAUUAUUAUACC--AG---------AGA-----------CCUCAAGAUGGCGCCAUAAAAACGCCAUUACUGUCUCCACCAGA--------- --------------..........................--.(---------(((-----------(.....(((((((.........)))))))...))))).......--------- ( -14.40, z-score = -4.32, R) >dp4.chr4_group3 8856301 114 - 11692001 ------AAAAAGAAACGGCAGAGGAAAAUAUUAUUACAACAAAACAACGCGGCAAAAGUUCAGCGUGCCGCAAGAUGGUGCCAUAAAAACGCCAUUACUGUCCCUACCACACACACAGAU ------..........((.((.(((.......................((((((...(.....).))))))..(((((((.........)))))))....))))).))............ ( -26.30, z-score = -2.64, R) >droPer1.super_8 38511 112 - 3966273 ------AAAAAGAAACGGCAGAGGAAAAUAUUAUUACAACAAAACAACGCGGCAAAAGUUCAGCGUGCCGCAAGAUGGUGCCAUAAAAACGCCAUUACUGUCCCUACCACACACAGAU-- ------..........((.((.(((.......................((((((...(.....).))))))..(((((((.........)))))))....))))).))..........-- ( -26.30, z-score = -2.62, R) >consensus ______GCGGAGAAAAGAAUAACGAAAAUGUUGUUACAAC_AAACAGCUCGGCAGA___________CCUCAAGAUGGCGCCGUAAAAACGCCAUUACUGUCUCCGCCGUGGA_______ ..................................................(((((.............(....)((((((.........))))))..))))).................. ( -8.96 = -9.40 + 0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:10:04 2011