| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 80,753 – 80,865 |
| Length | 112 |
| Max. P | 0.995143 |

| Location | 80,753 – 80,865 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 72.39 |
| Shannon entropy | 0.47504 |
| G+C content | 0.52521 |
| Mean single sequence MFE | -40.30 |
| Consensus MFE | -23.28 |
| Energy contribution | -22.08 |
| Covariance contribution | -1.20 |
| Combinations/Pair | 1.55 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.995143 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2RHet 80753 112 + 3288761 GUCAGGCCAUACGACCACAUGUCGCUGAGAGCUUGUUGUUA-CCGAACAACAGGUUUGAUGUGG----UCUUCUGGCCUUUACAACGUGUUCGGU--GUAGCAUACGAACAGGGAUGAC (((((((((...((((((((........((((((((((((.-...)))))))))))).))))))----))...))))).......(.((((((..--........)))))).)..)))) ( -44.40, z-score = -3.58, R) >droSim1.chrU 13678293 116 - 15797150 GUCAGGCCAUACGCCCACAUAUCGCUGGCAGCUCGCUGCAA-CCGAGCAACAAGUCCGAUGCGGCCCUUUUUUCGGCUUUCACAACGUGUUCGGU--GUGACGUGCAAACAGUGUCGGG ....(((.....)))..(....(((((...((.((..(((.-(((((((....((..((...((((........)))).))...)).))))))))--))..)).))...)))))...). ( -33.70, z-score = 0.97, R) >droYak2.chrU 26149408 112 - 28119190 GUCAGGCCAUACGCCCACUUGUCGCUGAGAUCUUGUUGCUA-CCGAACAACAGGUUCGAUGUGG----UCUUCUGACCUCAACAACGUGUUCGGU--GUAGCAUACGAACAGCGAUGAC (((.(((.....))).....(((((((......((((((.(-(((((((....(((.(.((.((----((....)))).)).)))).))))))))--))))))......)))))))))) ( -42.80, z-score = -3.34, R) >droEre2.scaffold_4845 21332981 112 - 22589142 GUCAGGCCAUACGCCCACACAUCGCUGGGAGCUUGUUGCUA-CCGAACAACAAGUUCGACGUGG----UUUUCGAGCCUCAACAACGUGUUCGGU--GUAGCAUGCGAACAGCGAUGAC ....(((.....)))....((((((((...((.((((((.(-(((((((....(((.(..(.((----((....)))).)..)))).))))))))--)))))).))...)))))))).. ( -45.80, z-score = -3.60, R) >droVir3.scaffold_13324 464971 107 + 2960039 --------AUUCAAAGACACAUCGCUCUAGGCGCGUAACAAACCAAACAACUUGUCCGAUGCGG----UCUUUUGGCCGCAGCAAAGUGUUCGGCUCAUAACGUUCAAACAGCGAUGUC --------..........((((((((....(.((((......((.((((..((((....(((((----((....)))))))))))..)))).))......)))).)....)))))))). ( -34.80, z-score = -3.29, R) >consensus GUCAGGCCAUACGCCCACAUAUCGCUGAGAGCUUGUUGCUA_CCGAACAACAAGUUCGAUGUGG____UCUUCUGGCCUCAACAACGUGUUCGGU__GUAGCAUACGAACAGCGAUGAC ...................((((((((......((((((...(((((((..(((((.((.(........).)).)))))........)))))))...))))))......)))))))).. (-23.28 = -22.08 + -1.20)

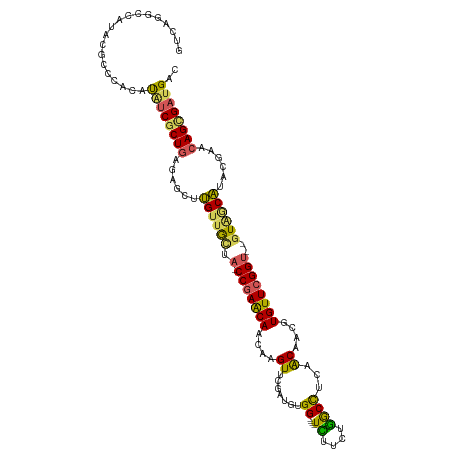
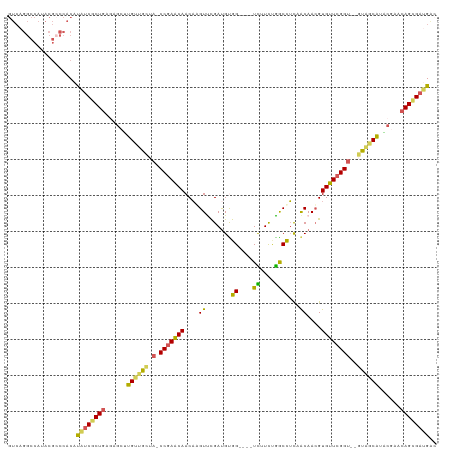
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:58:52 2011