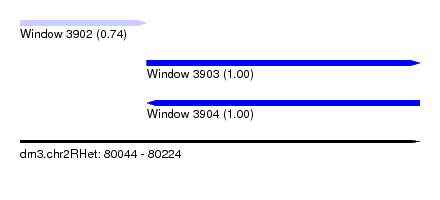
| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 80,044 – 80,224 |
| Length | 180 |
| Max. P | 0.998386 |

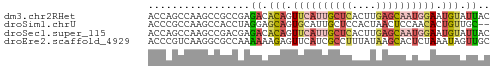
| Location | 80,044 – 80,101 |
|---|---|
| Length | 57 |
| Sequences | 4 |
| Columns | 57 |
| Reading direction | forward |
| Mean pairwise identity | 63.45 |
| Shannon entropy | 0.60806 |
| G+C content | 0.49179 |
| Mean single sequence MFE | -12.42 |
| Consensus MFE | -4.96 |
| Energy contribution | -7.02 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.744846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2RHet 80044 57 + 3288761 ACCAGCCAAGCCGCCGAGACACAGUUCAUUGCUCACUUGAGCAAUGGAAUGUAUUAC ....................(((.((((((((((....)))))))))).)))..... ( -16.20, z-score = -2.85, R) >droSim1.chrU 13677649 55 - 15797150 ACCCGCCAAGCCACCUAGGAGCAGUGCAUUGCUCCACUAACUCCAACACUGUUGC-- .................(((((((....)))))))........((((...)))).-- ( -10.70, z-score = -0.28, R) >droSec1.super_115 105658 57 + 121188 ACCAGCCAAGCCGACGAGACACAGUUCAUUGCUCACUUGAGCAAUGGAAUGUAUUAC ....................(((.((((((((((....)))))))))).)))..... ( -16.20, z-score = -3.15, R) >droEre2.scaffold_4929 2168159 57 - 26641161 ACCCGUCAAGGCGCCAAAAAAGAGUUCAUCGCCUUUAUAAGCACUCUAAAUAGUUGC ....((.((((((................)))))).))..(((((......)).))) ( -6.59, z-score = 0.24, R) >consensus ACCAGCCAAGCCGCCGAGAAACAGUUCAUUGCUCACUUAAGCAAUGGAAUGUAUUAC ....................(((.((((((((((....)))))))))).)))..... ( -4.96 = -7.02 + 2.06)
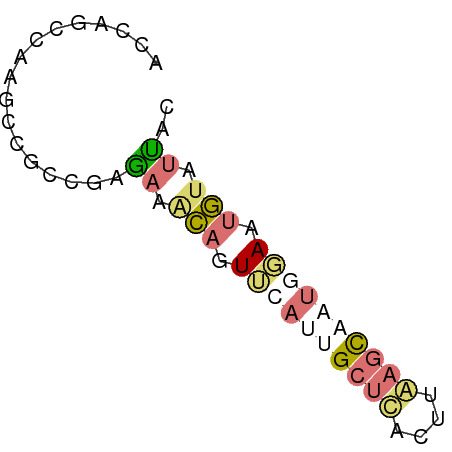
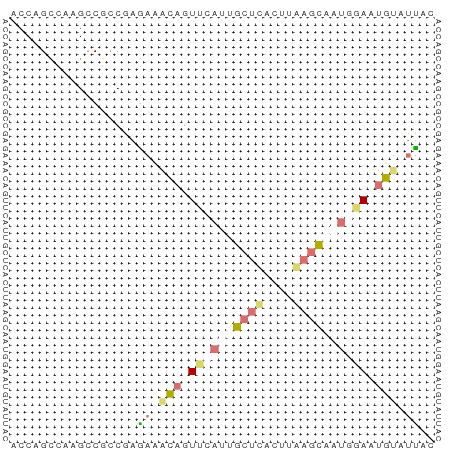
| Location | 80,101 – 80,224 |
|---|---|
| Length | 123 |
| Sequences | 6 |
| Columns | 123 |
| Reading direction | forward |
| Mean pairwise identity | 62.96 |
| Shannon entropy | 0.72307 |
| G+C content | 0.42951 |
| Mean single sequence MFE | -30.22 |
| Consensus MFE | -16.75 |
| Energy contribution | -17.95 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.26 |
| SVM RNA-class probability | 0.998113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
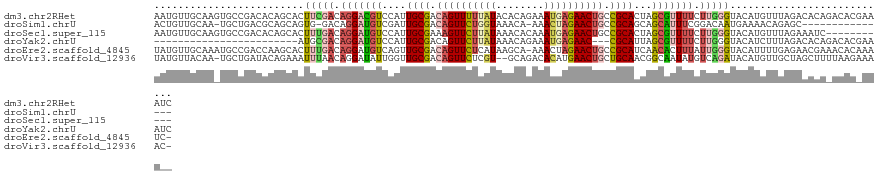
>dm3.chr2RHet 80101 123 + 3288761 AAUGUUGCAAGUGCCGACACAGCACUUCGACAGGACGUCCAUUGCGACAGUUUUUAUACACAGAAAUGAGAACUGCCGCACUAGCGUUUUCUUGGGUACAUGUUUAGACACAGACACGAAAUC ..(((((.((((((.......)))))))))))((((((....((((.((((((((((........)))))))))).))))...))))))..(((.((...(((......))).)).))).... ( -33.70, z-score = -1.94, R) >droSim1.chrU 13677704 105 - 15797150 ACUGUUGCAA-UGCUGACGCAGCAGUG-GACAGGAUGUCGAUUGCGACAGUUCUGGUAAACA-AAACUAGAACUGCCGCAGCAGCAUUUCGGACAAUGAAAACAGAGC--------------- .(((((...(-(((((..((.(((((.-(((.....))).)))))(.((((((((((.....-..)))))))))).)))..))))))((((.....)))))))))...--------------- ( -40.80, z-score = -4.22, R) >droSec1.super_115 105715 112 + 121188 AAUGUUGCAAGUGCCGACACAGCACUUUGACAGGAUGUCCAUUGCGAAAGUUCUUAUAAACACAAAUGAGAACUGCCGCACUAGCGUUUUCUUGGGUACAUGUUUAGAAAUC----------- ..(((((.((((((.......)))))))))))..((((((..((((..(((((((((........)))))))))..))))....((......)))).))))...........----------- ( -28.10, z-score = -0.91, R) >droYak2.chrU 26130691 96 - 28119190 ------------------------AUGCGACAGGAUGUCCAUUGCGACAGUUCUUAUAAACAGAAAUGAGAAC---CGCAUUAGCGUUUUCUUGGGUACAUCUUUAGACACAGACACGAAAUC ------------------------.(((...(((((((((.........((((((((........))))))))---(((....)))........)).)))))))..).))............. ( -19.10, z-score = -0.49, R) >droEre2.scaffold_4845 21332324 121 - 22589142 UAUGUUGCAAAUGCCGACCAAGCACUUUGACAGGAUGUCAGUUGCGACAGUUCUCAUAAGCA-AAACUAGAACUGCCGCAUCAACACUUUAUUGGGUACAUUUUGAGAACGAAACACAAAUC- ..((((((((.(((.......)))..(((((.....)))))))))))))(((((((..((..-...)).....((((.((............)))))).....)))))))............- ( -25.30, z-score = -0.39, R) >droVir3.scaffold_12936 444484 119 + 632223 UAUGUUACAA-UGCUGAUACAGAAAUUUAACAGGAUAUUGGUUGCGACAGUUCUCGU--GCAGACACAUGAACUGCUGCAACGGCAAUAUGUCAGAUACAUGUUGCUAGCUUUUAAGAAAAC- .......(((-(((((.((.(....).)).)))..)))))((((((.((((((..((--(....)))..)))))).))))))((((((((((.....))))))))))...............- ( -34.30, z-score = -2.70, R) >consensus AAUGUUGCAA_UGCCGACACAGCACUUUGACAGGAUGUCCAUUGCGACAGUUCUUAUAAACAGAAACGAGAACUGCCGCACCAGCAUUUUCUUGGGUACAUGUUGAGAAACA_ACAC_AAA__ .........................(((((.(((((((....((((.((((((((((........)))))))))).))))...))))))).)))))........................... (-16.75 = -17.95 + 1.20)

| Location | 80,101 – 80,224 |
|---|---|
| Length | 123 |
| Sequences | 6 |
| Columns | 123 |
| Reading direction | reverse |
| Mean pairwise identity | 62.96 |
| Shannon entropy | 0.72307 |
| G+C content | 0.42951 |
| Mean single sequence MFE | -30.69 |
| Consensus MFE | -16.87 |
| Energy contribution | -17.32 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.998386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
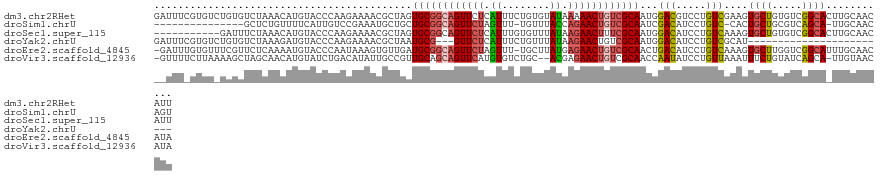
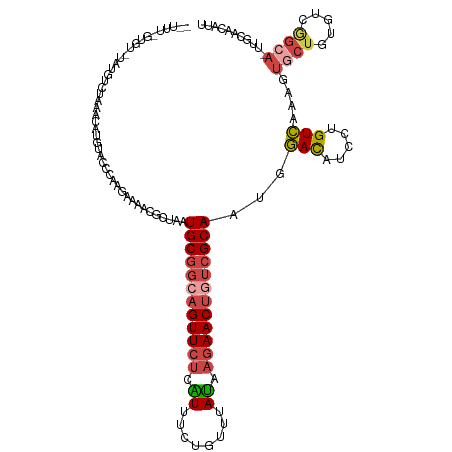
>dm3.chr2RHet 80101 123 - 3288761 GAUUUCGUGUCUGUGUCUAAACAUGUACCCAAGAAAACGCUAGUGCGGCAGUUCUCAUUUCUGUGUAUAAAAACUGUCGCAAUGGACGUCCUGUCGAAGUGCUGUGUCGGCACUUGCAACAUU .......(((..(((((...(((.((((....((..((((((.((((((((((..(((....)))......)))))))))).))).)))....))...)))))))...)))))..)))..... ( -35.90, z-score = -1.98, R) >droSim1.chrU 13677704 105 + 15797150 ---------------GCUCUGUUUUCAUUGUCCGAAAUGCUGCUGCGGCAGUUCUAGUUU-UGUUUACCAGAACUGUCGCAAUCGACAUCCUGUC-CACUGCUGCGUCAGCA-UUGCAACAGU ---------------...........(((((....(((((((.(((((((((...(((((-((.....)))))))((((....))))........-.))))))))).)))))-))...))))) ( -35.70, z-score = -3.20, R) >droSec1.super_115 105715 112 - 121188 -----------GAUUUCUAAACAUGUACCCAAGAAAACGCUAGUGCGGCAGUUCUCAUUUGUGUUUAUAAGAACUUUCGCAAUGGACAUCCUGUCAAAGUGCUGUGUCGGCACUUGCAACAUU -----------..(((((.............)))))..((...(((((.((((((.((........)).)))))).)))))...(((.....))).((((((((...))))))))))...... ( -26.22, z-score = -0.48, R) >droYak2.chrU 26130691 96 + 28119190 GAUUUCGUGUCUGUGUCUAAAGAUGUACCCAAGAAAACGCUAAUGCG---GUUCUCAUUUCUGUUUAUAAGAACUGUCGCAAUGGACAUCCUGUCGCAU------------------------ (((...(((((((((((....)))..............((....(((---(((((.((........)).)))))))).)).))))))))...)))....------------------------ ( -19.90, z-score = -0.62, R) >droEre2.scaffold_4845 21332324 121 + 22589142 -GAUUUGUGUUUCGUUCUCAAAAUGUACCCAAUAAAGUGUUGAUGCGGCAGUUCUAGUUU-UGCUUAUGAGAACUGUCGCAACUGACAUCCUGUCAAAGUGCUUGGUCGGCAUUUGCAACAUA -...(((.((..((((.....)))).)).)))....((((((.((((((((((((.((..-.....)).))))))))))))..((((.....))))(((((((.....))))))).)))))). ( -34.50, z-score = -2.01, R) >droVir3.scaffold_12936 444484 119 - 632223 -GUUUUCUUAAAAGCUAGCAACAUGUAUCUGACAUAUUGCCGUUGCAGCAGUUCAUGUGUCUGC--ACGAGAACUGUCGCAACCAAUAUCCUGUUAAAUUUCUGUAUCAGCA-UUGUAACAUA -(((((....)))))..((((.((((.....)))).)))).(((((.(((((((.((((....)--))).))))))).)))))........(((((.....(((...)))..-...))))).. ( -31.90, z-score = -2.62, R) >consensus __UUU_GUGU_UAUGUCUAAACAUGUACCCAAGAAAACGCUAAUGCGGCAGUUCUCAUUUCUGUUUAUAAGAACUGUCGCAAUGGACAUCCUGUCAAAGUGCUGUGUCGGCA_UUGCAACAUU ...........................................((((((((((((.((........)).))))))))))))...(((.....)))....((((.....))))........... (-16.87 = -17.32 + 0.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:58:51 2011