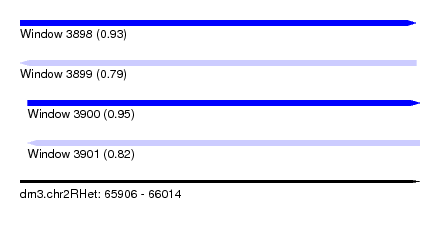
| Sequence ID | dm3.chr2RHet |
|---|---|
| Location | 65,906 – 66,014 |
| Length | 108 |
| Max. P | 0.950312 |

| Location | 65,906 – 66,013 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 66.86 |
| Shannon entropy | 0.59036 |
| G+C content | 0.43277 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -12.36 |
| Energy contribution | -13.72 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.930741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2RHet 65906 107 + 3288761 GGAGUUGGCACUGACGCUAGCAGAAGCAGCGACGGUGCUGGGGGGUAUAAAUACCACCCAAUUUUGUACUUUAAAUUUAGAUUCUAAAUAUCUUCUCGUGAAAACAA (((((((((((((.((((.((....)))))).)))))))(((.((((....)))).)))....................))))))............((....)).. ( -33.20, z-score = -3.13, R) >droSim1.chr3h_random 391462 94 - 1452968 GGAGUUGGCACUGAAGCUAGCAGAAGCAGCGACGGUGCU-------AUAAAUACCACCCAAUUUUGUACUUUAAAUUUAGUUCCGA------UUUUUGUCAAAAUAA ((.((((((((((..(((.((....)))))..)))))))-------)...)).)).....((((((.((...(((((.......))------)))..)))))))).. ( -22.70, z-score = -1.73, R) >droSec1.super_134 19166 101 - 53330 UGAGUUGGCACUGACGCUAGCAGAAGCAGCGACGGUGCUGGGGGGUAUAAAUACCACCCAAUUUUGUACUUUAAAUUUAGUUCCGA------UUUUUGUCAAAAUAA ......(((((((.((((.((....)))))).)))))))(((.((((....)))).))).((((((.((...(((((.......))------)))..)))))))).. ( -31.80, z-score = -2.82, R) >droEre2.scaffold_4929 760939 99 - 26641161 GGAGUUGGCACCGACGCUGGCAGAGGCAGCGACGGAGCUGGG--GCAUAAAUAUCACCCAAACUUGUAAUUCAGAUUUAUUUCCAA------UUUUUAUGCAAAUAA ((((..(((.(((.(((((.(...).))))).))).)))(((--............))).....................))))..------............... ( -24.50, z-score = -0.72, R) >droAna3.scaffold_13417 745187 96 - 6960332 -GACAUCG-AGGGACGCCAGCAGCAGCAGCAACCACGACCGAGAUCGCGGCAGCGAUGGAGCAACGCA---UAGGUGUAAUUAUUAA-----UUAUAAGAAACAUA- -.(((((.-.((....)).((.((....))..(((((.(((......)))...)).)))......)).---..))))).........-----..............- ( -17.80, z-score = 0.50, R) >consensus GGAGUUGGCACUGACGCUAGCAGAAGCAGCGACGGUGCUGGGG_GCAUAAAUACCACCCAAUUUUGUACUUUAAAUUUAGUUCCGA______UUUUUGUCAAAAUAA ......(((((((.((((.((....)))))).))))))).................................................................... (-12.36 = -13.72 + 1.36)

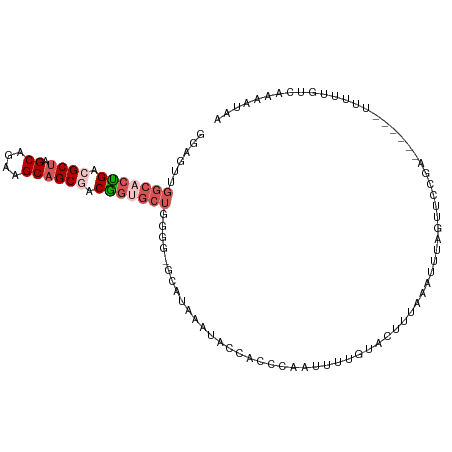
| Location | 65,906 – 66,013 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 66.86 |
| Shannon entropy | 0.59036 |
| G+C content | 0.43277 |
| Mean single sequence MFE | -24.44 |
| Consensus MFE | -9.78 |
| Energy contribution | -10.38 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.787897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2RHet 65906 107 - 3288761 UUGUUUUCACGAGAAGAUAUUUAGAAUCUAAAUUUAAAGUACAAAAUUGGGUGGUAUUUAUACCCCCCAGCACCGUCGCUGCUUCUGCUAGCGUCAGUGCCAACUCC .(((((((....)))))))..................(((.......((((.((((....)))).))))((((.(.((((((....)).)))).).))))..))).. ( -29.10, z-score = -3.07, R) >droSim1.chr3h_random 391462 94 + 1452968 UUAUUUUGACAAAAA------UCGGAACUAAAUUUAAAGUACAAAAUUGGGUGGUAUUUAU-------AGCACCGUCGCUGCUUCUGCUAGCUUCAGUGCCAACUCC ...((((((......------))))))...........((((.......((((.((....)-------).))))(..(((((....)).)))..).))))....... ( -17.00, z-score = -0.03, R) >droSec1.super_134 19166 101 + 53330 UUAUUUUGACAAAAA------UCGGAACUAAAUUUAAAGUACAAAAUUGGGUGGUAUUUAUACCCCCCAGCACCGUCGCUGCUUCUGCUAGCGUCAGUGCCAACUCA ...((((((......------))))))..........(((.......((((.((((....)))).))))((((.(.((((((....)).)))).).))))..))).. ( -26.80, z-score = -2.61, R) >droEre2.scaffold_4929 760939 99 + 26641161 UUAUUUGCAUAAAAA------UUGGAAAUAAAUCUGAAUUACAAGUUUGGGUGAUAUUUAUGC--CCCAGCUCCGUCGCUGCCUCUGCCAGCGUCGGUGCCAACUCC ......(((((((..------(..((......))..)(((((........))))).)))))))--....((.(((.(((((.......))))).))).))....... ( -25.70, z-score = -1.69, R) >droAna3.scaffold_13417 745187 96 + 6960332 -UAUGUUUCUUAUAA-----UUAAUAAUUACACCUA---UGCGUUGCUCCAUCGCUGCCGCGAUCUCGGUCGUGGUUGCUGCUGCUGCUGGCGUCCCU-CGAUGUC- -..............-----................---...((.((..((..((.((((((((....)))))))).))))..)).)).((((((...-.))))))- ( -23.60, z-score = -0.83, R) >consensus UUAUUUUCACAAAAA______UAGGAACUAAAUUUAAAGUACAAAAUUGGGUGGUAUUUAUAC_CCCCAGCACCGUCGCUGCUUCUGCUAGCGUCAGUGCCAACUCC .....................................................................((((.(.((((((....)).)))).).))))....... ( -9.78 = -10.38 + 0.60)

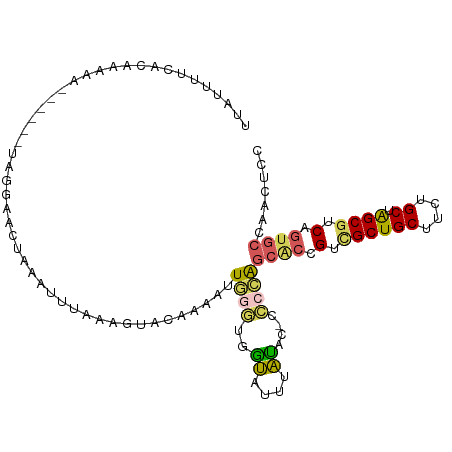
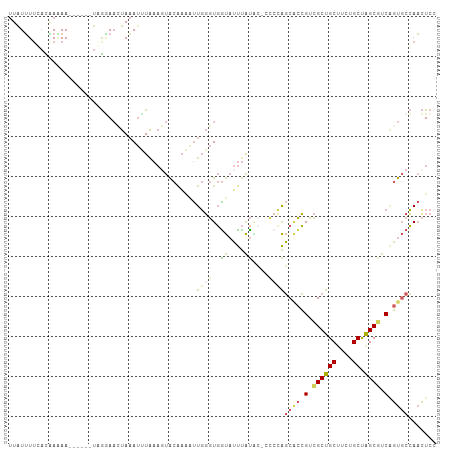
| Location | 65,908 – 66,014 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 66.44 |
| Shannon entropy | 0.59662 |
| G+C content | 0.42091 |
| Mean single sequence MFE | -25.27 |
| Consensus MFE | -12.30 |
| Energy contribution | -13.38 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.950312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2RHet 65908 106 + 3288761 AGUUGGCACUGACGCUAGCAGAAGCAGCGACGGUGCUGGGGGGUAUAAAUACCACCCAAUUUUGUACUUUAAAUUUAGAUUCUAAAUAUCUUCUCGUGAAAACAAA ....(((((((.((((.((....)))))).)))))))(((.((((....)))).)))...(((((..((((.....((((.......)))).....)))).))))) ( -31.50, z-score = -2.92, R) >droSim1.chr3h_random 391464 93 - 1452968 AGUUGGCACUGAAGCUAGCAGAAGCAGCGACGGUGCU-------AUAAAUACCACCCAAUUUUGUACUUUAAAUUUAGUUCCGA------UUUUUGUCAAAAUAAA ...((((((((..(((.((....)))))..)))))))-------).............((((((.((...(((((.......))------)))..))))))))... ( -21.90, z-score = -1.97, R) >droSec1.super_134 19168 100 - 53330 AGUUGGCACUGACGCUAGCAGAAGCAGCGACGGUGCUGGGGGGUAUAAAUACCACCCAAUUUUGUACUUUAAAUUUAGUUCCGA------UUUUUGUCAAAAUAAA ....(((((((.((((.((....)))))).)))))))(((.((((....)))).))).((((((.((...(((((.......))------)))..))))))))... ( -31.80, z-score = -3.00, R) >droEre2.scaffold_4929 760941 98 - 26641161 AGUUGGCACCGACGCUGGCAGAGGCAGCGACGGAGCUGGG--GCAUAAAUAUCACCCAAACUUGUAAUUCAGAUUUAUUUCCAA------UUUUUAUGCAAAUAAA ..(..((.(((.(((((.(...).))))).))).))..).--(((((((........(((.(((.....))).)))........------..)))))))....... ( -23.57, z-score = -0.74, R) >droAna3.scaffold_13417 745189 95 - 6960332 CAUCGAGG--GACGCCAGCAGCAGCAGCAACCACGACCGAGAUCGCGGCAGCGAUGGAGCAACGCA---UAGGUGUAAUUAUUAA-----UUAUAAGAAACAUAA- .....((.--.(((((.((....)).((..(((((.(((......)))...)).))).))......---..)))))..)).....-----...............- ( -17.60, z-score = 0.56, R) >consensus AGUUGGCACUGACGCUAGCAGAAGCAGCGACGGUGCUGGGG_GCAUAAAUACCACCCAAUUUUGUACUUUAAAUUUAGUUCCGA______UUUUUGUCAAAAUAAA ....(((((((.((((.((....)))))).)))))))..................................................................... (-12.30 = -13.38 + 1.08)

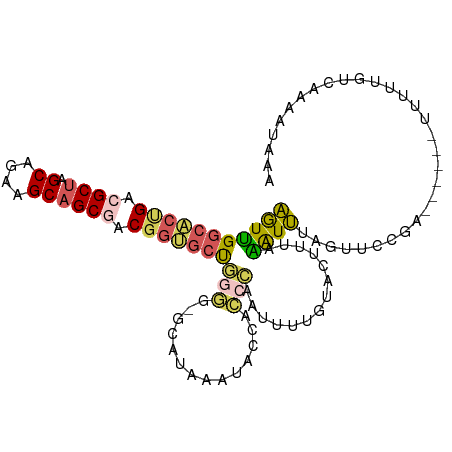
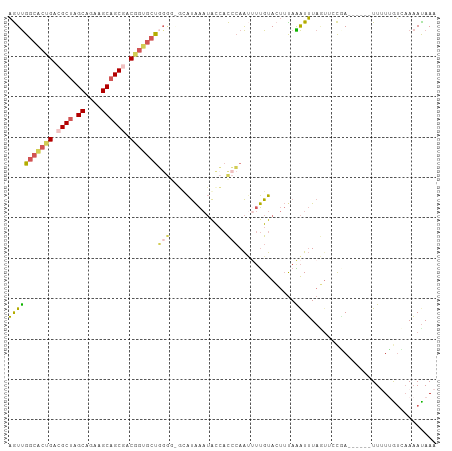
| Location | 65,908 – 66,014 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 66.44 |
| Shannon entropy | 0.59662 |
| G+C content | 0.42091 |
| Mean single sequence MFE | -24.22 |
| Consensus MFE | -9.90 |
| Energy contribution | -10.38 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.820003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2RHet 65908 106 - 3288761 UUUGUUUUCACGAGAAGAUAUUUAGAAUCUAAAUUUAAAGUACAAAAUUGGGUGGUAUUUAUACCCCCCAGCACCGUCGCUGCUUCUGCUAGCGUCAGUGCCAACU ..(((((((....)))))))............................((((.((((....)))).))))((((.(.((((((....)).)))).).))))..... ( -29.00, z-score = -2.99, R) >droSim1.chr3h_random 391464 93 + 1452968 UUUAUUUUGACAAAAA------UCGGAACUAAAUUUAAAGUACAAAAUUGGGUGGUAUUUAU-------AGCACCGUCGCUGCUUCUGCUAGCUUCAGUGCCAACU ((((((((((......------)))))).))))......((((.......((((.((....)-------).))))(..(((((....)).)))..).))))..... ( -17.20, z-score = -0.14, R) >droSec1.super_134 19168 100 + 53330 UUUAUUUUGACAAAAA------UCGGAACUAAAUUUAAAGUACAAAAUUGGGUGGUAUUUAUACCCCCCAGCACCGUCGCUGCUUCUGCUAGCGUCAGUGCCAACU ((((((((((......------)))))).))))...............((((.((((....)))).))))((((.(.((((((....)).)))).).))))..... ( -26.90, z-score = -2.69, R) >droEre2.scaffold_4929 760941 98 + 26641161 UUUAUUUGCAUAAAAA------UUGGAAAUAAAUCUGAAUUACAAGUUUGGGUGAUAUUUAUGC--CCCAGCUCCGUCGCUGCCUCUGCCAGCGUCGGUGCCAACU .......(((((((..------(..((......))..)(((((........))))).)))))))--....((.(((.(((((.......))))).))).))..... ( -25.70, z-score = -1.67, R) >droAna3.scaffold_13417 745189 95 + 6960332 -UUAUGUUUCUUAUAA-----UUAAUAAUUACACCUA---UGCGUUGCUCCAUCGCUGCCGCGAUCUCGGUCGUGGUUGCUGCUGCUGCUGGCGUC--CCUCGAUG -...............-----................---.(((((((..((.(((.((((((((....)))))))).)).).))..)).))))).--........ ( -22.30, z-score = -0.60, R) >consensus UUUAUUUUCACAAAAA______UAGGAACUAAAUUUAAAGUACAAAAUUGGGUGGUAUUUAUAC_CCCCAGCACCGUCGCUGCUUCUGCUAGCGUCAGUGCCAACU ......................................................................((((.(.((((((....)).)))).).))))..... ( -9.90 = -10.38 + 0.48)

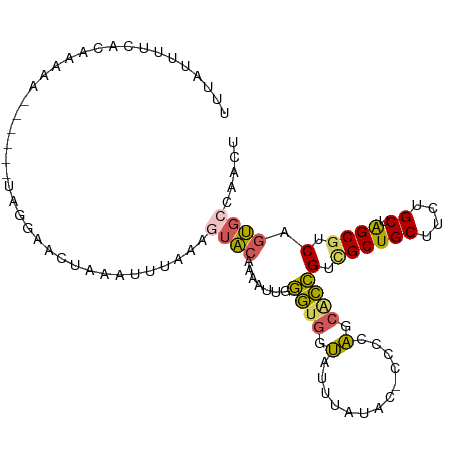
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:58:49 2011