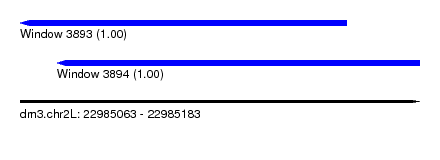
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 22,985,063 – 22,985,183 |
| Length | 120 |
| Max. P | 0.999442 |

| Location | 22,985,063 – 22,985,161 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 60.30 |
| Shannon entropy | 0.56562 |
| G+C content | 0.55255 |
| Mean single sequence MFE | -45.53 |
| Consensus MFE | -25.15 |
| Energy contribution | -27.40 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.89 |
| SVM RNA-class probability | 0.999442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 22985063 98 - 23011544 -GACUGGGGCUUCAGUCCUCGAGCCAGGCGGACCAUAUAUGACGGACUCAUGGCACCUUGUGUGCUGUUUGGUGCCCCGGUAUGGAACAA-GUAGCUGCC------------ -.((((((((.((((((((((.....((....)).....))).))))..(((((((.....))))))).))).)))))))).........-.........------------ ( -31.00, z-score = 0.77, R) >droVir3.scaffold_9500 4936 112 + 5668 UGACUGGGGCCUAAGCAGACGUGCUGUCCGCACCAUCUAUGAGGGUCUGUUUGUUGCAUGUGCAGCAUAUGGAUCGUCUGUAUGGUGCAAGGCGGUCACGACUGUGGUCGGC .......((((...((((.(((((((((.(((((((.((.((.(((((((.((((((....)))))).))))))).)))).)))))))..))))).)))).))))))))... ( -51.80, z-score = -3.44, R) >droGri2.scaffold_15040 117682 111 - 162499 -GACUGGGGCCUCAGCAGAACUGCUGCUCGCGCCGUAUACGAUGGUCUAUUUGUUGCCUGUGCAACUUAUGGAUCGUCGGUAUGGUGUAAAGUAGUUUCGACUGUUGUAGGC -.......((((((((((....((((((.(((((((((.(((((((((((..(((((....)))))..))))))))))))))))))))..)))))).....)))))).)))) ( -53.80, z-score = -6.25, R) >consensus _GACUGGGGCCUCAGCAGACGUGCUGCCCGCACCAUAUAUGACGGUCUAUUUGUUGCAUGUGCAACAUAUGGAUCGUCGGUAUGGUGCAA_GUAGUUACGACUGU_GU_GGC .(((((((((...((((....))))))))(((((((((.(((.(((((((..(((((....)))))..))))))).))))))))))))....)))))............... (-25.15 = -27.40 + 2.25)

| Location | 22,985,074 – 22,985,183 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 55.56 |
| Shannon entropy | 0.64008 |
| G+C content | 0.55802 |
| Mean single sequence MFE | -43.11 |
| Consensus MFE | -20.79 |
| Energy contribution | -23.37 |
| Covariance contribution | 2.57 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.31 |
| SVM RNA-class probability | 0.998277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 22985074 109 - 23011544 ---GCUGGCGCGUGUGCUUCGAGCCGACUGGGGCUUCAGUCCUCGAGCCAGGCGGACCAUAUAUGACGGACUCAUGGCACCUUGUGUGCUGUUUGGUGCCCCGGUAUGGAAC ---.....(((.((.(((.((((..((((((....))))))))))))))).)))..((((((((((.....))))((((((.............))))))...))))))... ( -40.52, z-score = 0.12, R) >droVir3.scaffold_9500 4960 112 + 5668 GCAAGUGCGUCGUAUUUUGAGGUCUGACUGGGGCCUAAGCAGACGUGCUGUCCGCACCAUCUAUGAGGGUCUGUUUGUUGCAUGUGCAGCAUAUGGAUCGUCUGUAUGGUGC ...((..((((........((((((.....)))))).....))))..))....(((((((.((.((.(((((((.((((((....)))))).))))))).)))).))))))) ( -47.32, z-score = -3.18, R) >droGri2.scaffold_15040 117706 92 - 162499 --------------------GAAGUGACUGGGGCCUCAGCAGAACUGCUGCUCGCGCCGUAUACGAUGGUCUAUUUGUUGCCUGUGCAACUUAUGGAUCGUCGGUAUGGUGU --------------------..........((((...((((....))))))))(((((((((.(((((((((((..(((((....)))))..)))))))))))))))))))) ( -41.50, z-score = -4.30, R) >consensus _____UG___CGU_U__U__GAACUGACUGGGGCCUCAGCAGACGUGCUGCCCGCACCAUAUAUGACGGUCUAUUUGUUGCAUGUGCAACAUAUGGAUCGUCGGUAUGGUGC ..............................((((...((((....))))))))(((((((((.(((.(((((((..(((((....)))))..))))))).)))))))))))) (-20.79 = -23.37 + 2.57)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:58:44 2011