| Sequence ID | dm3.chr2L |
|---|---|
| Location | 22,923,871 – 22,923,926 |
| Length | 55 |
| Max. P | 0.827229 |

| Location | 22,923,871 – 22,923,926 |
|---|---|
| Length | 55 |
| Sequences | 5 |
| Columns | 57 |
| Reading direction | forward |
| Mean pairwise identity | 76.30 |
| Shannon entropy | 0.41010 |
| G+C content | 0.53799 |
| Mean single sequence MFE | -16.32 |
| Consensus MFE | -11.50 |
| Energy contribution | -12.42 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.800864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
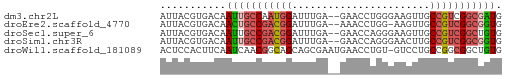
>dm3.chr2L 22923871 55 + 23011544 AUUACGUGACAAUUGCCAAUGGAUUUGA--GAACCUGGGAAGUUGCCGUCGGCGAUG ...........((((((.((((.(((.(--(...)).))).....)))).)))))). ( -11.00, z-score = 0.13, R) >droEre2.scaffold_4770 14631890 54 - 17746568 AUUACGUGACAACUGCCGACGGAUUUGA--AAACCUGG-AAGUUGCCGUCGGCGGUG ...........(((((((((((.(((.(--.....).)-))....))))))))))). ( -21.30, z-score = -3.39, R) >droSec1.super_6 1406100 55 - 4358794 AUUACGUGACAAUUGCCGACGGAUUUGA--GAACCAGGGAAGUUGCCGUCGGCUGUG .........((...((((((((.((((.--....)))).......))))))))..)) ( -16.10, z-score = -1.36, R) >droSim1.chr3R 24615378 55 - 27517382 AUUACGUGACAAUUGCCGACGGAUUUGA--GAACCAGGGAACUUGCCGUCGGCGGUG ...........(((((((((((....((--(..(....)..))).))))))))))). ( -19.50, z-score = -2.45, R) >droWil1.scaffold_181089 81907 56 - 12369635 ACUCCACUUCAAUCAACGGCAGCAGCGAAUGAACCUGU-GUCCUGCCGGCGGCUGUG .........((..(..((((((..(((........)))-...))))))..)..)).. ( -13.70, z-score = 0.32, R) >consensus AUUACGUGACAAUUGCCGACGGAUUUGA__GAACCUGGGAAGUUGCCGUCGGCGGUG ...........(((((((((((.......................))))))))))). (-11.50 = -12.42 + 0.92)
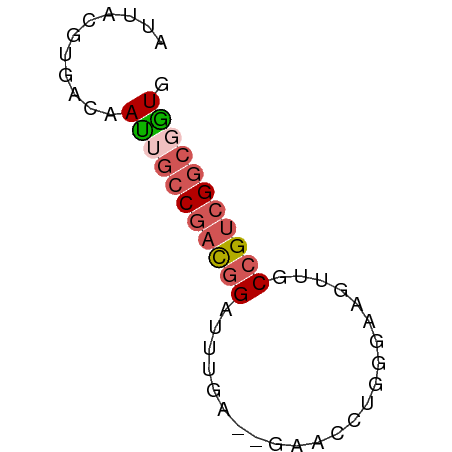

| Location | 22,923,871 – 22,923,926 |
|---|---|
| Length | 55 |
| Sequences | 5 |
| Columns | 57 |
| Reading direction | reverse |
| Mean pairwise identity | 76.30 |
| Shannon entropy | 0.41010 |
| G+C content | 0.53799 |
| Mean single sequence MFE | -13.92 |
| Consensus MFE | -10.40 |
| Energy contribution | -10.52 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.827229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
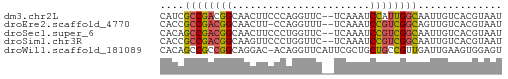
>dm3.chr2L 22923871 55 - 23011544 CAUCGCCGACGGCAACUUCCCAGGUUC--UCAAAUCCAUUGGCAAUUGUCACGUAAU ....(((((.(....)......((...--......)).))))).............. ( -8.20, z-score = 0.14, R) >droEre2.scaffold_4770 14631890 54 + 17746568 CACCGCCGACGGCAACUU-CCAGGUUU--UCAAAUCCGUCGGCAGUUGUCACGUAAU ....((((((((.((((.-...)))).--......)))))))).............. ( -15.60, z-score = -2.34, R) >droSec1.super_6 1406100 55 + 4358794 CACAGCCGACGGCAACUUCCCUGGUUC--UCAAAUCCGUCGGCAAUUGUCACGUAAU ....((((((((.((((.....)))).--......)))))))).............. ( -15.70, z-score = -2.64, R) >droSim1.chr3R 24615378 55 + 27517382 CACCGCCGACGGCAAGUUCCCUGGUUC--UCAAAUCCGUCGGCAAUUGUCACGUAAU ....((((((((..((..(....)..)--).....)))))))).............. ( -16.90, z-score = -2.63, R) >droWil1.scaffold_181089 81907 56 + 12369635 CACAGCCGCCGGCAGGAC-ACAGGUUCAUUCGCUGCUGCCGUUGAUUGAAGUGGAGU .....((((((((((...-.((((......).))))))))).........))))... ( -13.20, z-score = 1.21, R) >consensus CACCGCCGACGGCAACUUCCCAGGUUC__UCAAAUCCGUCGGCAAUUGUCACGUAAU ....((((((((.......................)))))))).............. (-10.40 = -10.52 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:58:40 2011