| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,049,815 – 2,049,884 |
| Length | 69 |
| Max. P | 0.641616 |

| Location | 2,049,815 – 2,049,884 |
|---|---|
| Length | 69 |
| Sequences | 10 |
| Columns | 84 |
| Reading direction | forward |
| Mean pairwise identity | 73.02 |
| Shannon entropy | 0.49320 |
| G+C content | 0.43446 |
| Mean single sequence MFE | -15.49 |
| Consensus MFE | -8.17 |
| Energy contribution | -8.73 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.641616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2049815 69 + 23011544 -----------CCCCAAUCCAGUCGAGUCAUGCGAUUGUAAUAUUUAUGCAGUUUG----UGCAACGUUCGCUAUAUUUGACAG -----------..........(((((((...((((.(((........((((.....----))))))).))))...))))))).. ( -15.80, z-score = -2.20, R) >droEre2.scaffold_4929 2097678 68 + 26641161 ------------GCCGAUCCAGUCGAGUCAUGCGAUUGCAAUAUUUAUGCAGUUAG----UGCAGCGUUCGCUAUAUUUGACAG ------------.........(((((((...((((.(((........((((.....----))))))).))))...))))))).. ( -17.50, z-score = -1.10, R) >droYak2.chr2L 2035508 80 + 22324452 CUUCCCCAUGAAGCCGAUCCAGUCGAGUCAUGCGAUUGCAAUAUUUAUGCAGUUAG----UGCAGCGUUCGCUAUAUUUGACAG ((((.....))))........(((((((...((((.(((........((((.....----))))))).))))...))))))).. ( -18.50, z-score = -1.24, R) >droSec1.super_14 1996518 69 + 2068291 -----------CCCCAAUCCAGUCGAGUCAUGCGAUUGCAAUAUUUAUGCAGUUUG----UGCAACGUUCGCUAUAUUUGACAG -----------..........(((((((...(((((((((..(((.....)))...----)))))...))))...))))))).. ( -16.30, z-score = -2.05, R) >droSim1.chr2L 2019722 69 + 22036055 -----------CCCCAAUCCAGUCGAGUCAUGCGAUUGCAAUAUUUAUGCAGUUUG----UGCAGCGUUCGCUAUAUUUGACAG -----------..........(((((((...((((.(((........((((.....----))))))).))))...))))))).. ( -17.50, z-score = -2.20, R) >droMoj3.scaffold_6473 7870357 57 - 16943266 -----------CCUCAAUGCAAUCGAGCUAUGCAGCUGUAAUAUCU-UAAGGUCUA------CUCAGUUUGGCAU--------- -----------.....((((...((((((..(.((((.(((....)-)).)).)).------)..))))))))))--------- ( -7.80, z-score = 1.06, R) >droWil1.scaffold_180708 11708686 63 - 12563649 ---------------------GUCGAGUCAUGCCGUUGCAAUAUUUAUGCAGUUUAACUAUGUUCGUUUGGCCAUGUUUGACAG ---------------------((((((.((((((((((((.......)))))...(((.......)))))).)))))))))).. ( -14.90, z-score = -1.35, R) >droPer1.super_8 26611 59 + 3966273 ---------------------GUCGAGUCAUGGCAUUGCAAUAUUUAUGCAGUUUG----UGCCACGUUCGCUAUAUUUGACAG ---------------------(((((((.(((((..((((.......))))(..((----.....))..))))))))))))).. ( -15.60, z-score = -1.54, R) >dp4.chr4_group3 8844297 59 + 11692001 ---------------------GUCGAGUCAUGGCAUUGCAAUAUUUAUGCAGUUUG----UGCCACGUUCGCUAUAUUUGACAG ---------------------(((((((.(((((..((((.......))))(..((----.....))..))))))))))))).. ( -15.60, z-score = -1.54, R) >droAna3.scaffold_12916 9764897 59 - 16180835 ---------------------GUCGAGUCAUGCCGUUGCAAUAUUUAUGCAGUUUG----UGCUGCGUUCGCUAUAUUUGACAG ---------------------(((((((..(((....)))......(((((((...----.))))))).......))))))).. ( -15.40, z-score = -1.02, R) >consensus _____________CC_AUCCAGUCGAGUCAUGCGAUUGCAAUAUUUAUGCAGUUUG____UGCAACGUUCGCUAUAUUUGACAG .....................(((((((...((((.((((.......))))((........)).....))))...))))))).. ( -8.17 = -8.73 + 0.56)

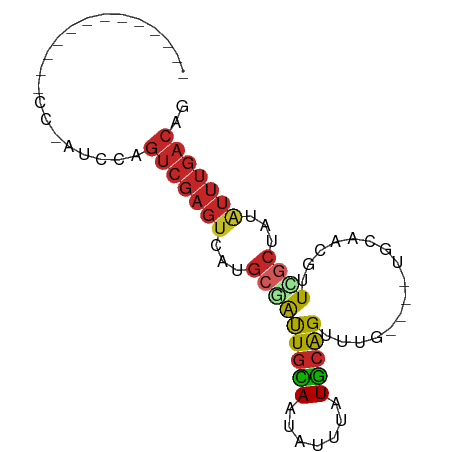
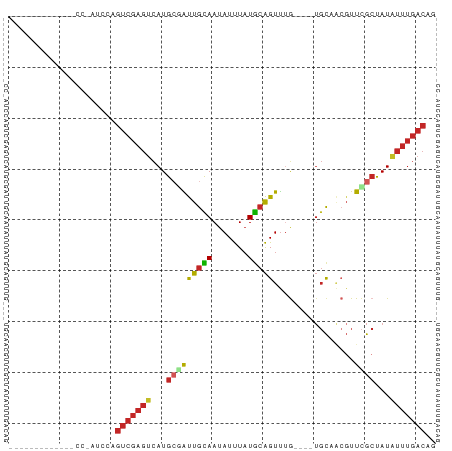
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:10:01 2011