| Sequence ID | dm3.chr2L |
|---|---|
| Location | 22,490,044 – 22,490,148 |
| Length | 104 |
| Max. P | 0.679084 |

| Location | 22,490,044 – 22,490,148 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 51.12 |
| Shannon entropy | 0.67011 |
| G+C content | 0.52726 |
| Mean single sequence MFE | -33.07 |
| Consensus MFE | -13.25 |
| Energy contribution | -12.71 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.594883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 22490044 104 + 23011544 GCUCCCUCAAUUCUUAUUAGGGUGGCUUUCCUCUUCAUUUCAA-UGACGAUAUGAAG--UGGCGUCACCUCCACUAGUACUUCUAGGGUUGCAGUGGGACAGGUACG ..((((.(...........(((((((...((.((((((.((..-....)).))))))--.)).)))))))((.((((.....)))))).....).))))........ ( -31.70, z-score = -1.78, R) >droYak2.chrX_random 1069736 99 - 1802292 ----CGUGGAUGUCUAGGGGAUCAACUCAAAUUAUUGUUUCGAGUGCUUUUGUGGGGGUUGACACCAGCGCCCCUGUUAUGCAGAGCAGUGCCUGCCGCUGGG---- ----.((.(((.((((.((((...((((((((....)))).))))..)))).)))).))).)).((((((...(((.....))).((((...)))))))))).---- ( -32.10, z-score = -0.04, R) >droPer1.super_278 18121 99 + 27293 ----CGUGAAUGUCCAGGGGGUCGAUUCCGAGUAGGGUUUCGAGUGCUUUUGUUGGGGUUGACCUCAACGCCCCUGUAAUACAGAGCAGAGCCUGUCGCUGAG---- ----.((((.....((((((.((((..((......))..))))........(((((((....))))))).)))))).....(((.((...)))))))))....---- ( -35.40, z-score = -1.41, R) >consensus ____CGUGAAUGUCUAGGGGGUCGACUCAAAUUAUCGUUUCGAGUGCUUUUGUGGGGGUUGACAUCAACGCCCCUGUUAUACAGAGCAGUGCCUGCCGCUGGG____ .....(((......((((((((((((((((((.(..((.......))..).)))))))))))).......))))))....((((.(.....))))))))........ (-13.25 = -12.71 + -0.54)

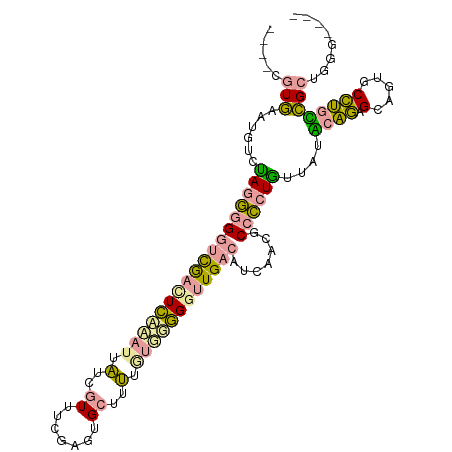
| Location | 22,490,044 – 22,490,148 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 51.12 |
| Shannon entropy | 0.67011 |
| G+C content | 0.52726 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -10.89 |
| Energy contribution | -9.46 |
| Covariance contribution | -1.42 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.679084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 22490044 104 - 23011544 CGUACCUGUCCCACUGCAACCCUAGAAGUACUAGUGGAGGUGACGCCA--CUUCAUAUCGUCA-UUGAAAUGAAGAGGAAAGCCACCCUAAUAAGAAUUGAGGGAGC ........((((.......((((((.....)))).)).((((.(.((.--((((((.(((...-.))).)))))).))...).))))..............)))).. ( -29.00, z-score = -2.02, R) >droYak2.chrX_random 1069736 99 + 1802292 ----CCCAGCGGCAGGCACUGCUCUGCAUAACAGGGGCGCUGGUGUCAACCCCCACAAAAGCACUCGAAACAAUAAUUUGAGUUGAUCCCCUAGACAUCCACG---- ----..(((.(((((...))))))))......(((((.(((.(((........)))...)))(((((((.......)))))))....)))))...........---- ( -25.60, z-score = -0.47, R) >droPer1.super_278 18121 99 - 27293 ----CUCAGCGACAGGCUCUGCUCUGUAUUACAGGGGCGUUGAGGUCAACCCCAACAAAAGCACUCGAAACCCUACUCGGAAUCGACCCCCUGGACAUUCACG---- ----.......((((((...)).))))....((((((.((((.((....)).))))........((((..((......))..)))).))))))..........---- ( -26.70, z-score = -1.00, R) >consensus ____CCCAGCGACAGGCACUGCUCUGAAUAACAGGGGCGCUGACGUCAACCCCCACAAAAGCACUCGAAACAAUAAUCAGAGUCGACCCCCUAGACAUUCACG____ .........(((((((......)))))....((((((.(((((.......................................)))))))))))........)).... (-10.89 = -9.46 + -1.42)

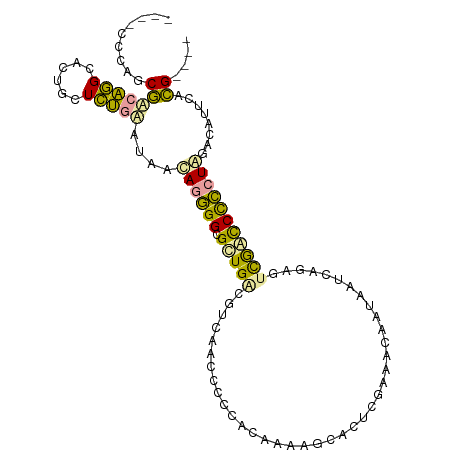
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:58:24 2011